Gene
KWMTBOMO08577
Annotation
PREDICTED:_uncharacterized_protein_LOC105386400_[Plutella_xylostella]
Transcription factor
Location in the cell
Nuclear Reliability : 2.944
Sequence
CDS
ATGCCGAGACGCTGTATATTAAAATGTGCACCAGAACCTGGTGTAAGGTTTCACGTATTTCCCAGCCTAGAGAGTAAACCTGAACGTTTTAAGGCATGGGTTCAAGCAATTGGAGGTAAACTGGATACCCCAGAGGACTATGAATACTACAGAAACCAACGCATATGTGATCGGCATTTTGCAAAAGAAAATAAAAACCGATACAATCGTGTGGACGCTTTGGGAGTTCCATCGCTTCATTTAATCGAAACTGATCTGGGAGAAAGAGAAAAAAATTCTTTAGGAGAAAGCTTTGCCTCCTCAGTACAAGAGACATTGAATTTGGATGTTATTGAACCTACAACTTCCAATGTCCTAGTTGAAGATATGATGACACGGAACAATAGTTTATCCTTGCGAAAATATAATATGTACCAAAGTCAAATCAAACAATTAAGGAAAAGAGTGAACACTTTTCAAAAAAGAGAAGATAACTTCAGGACAAGATTAGCAAATGCTGAAAAGATTACTGATGAATCTGCATTTAAAAGGATTGCCAAAAATATGACTCTGCCTGCAAAGATTTTTACGAATATGCAGTTTCGCACTACGTCAAAAAAACCGTCGGGCAGGAGATTTCTATTAGAAGAAAAAGTGCTATCATTAACACTCTACAAAAGAAGTCCCAAATGTTATTCAATGTTGTCCAGGTATTTTTCTTTACCAGCGCCTAAAACTTTGAAAAGACTTCTCAGTGAAATTAAATTGGGCCCTGGTATCAATACGATTGTTATGGAAAAATTAAAGAAAACTGTTAGTGAGCTCACCATGGAAGACCGCTTGTGCACTTTAATTTTTGATGAGATGTCTATAACCCCTCAGCTGCATTATGATGCAGCAAAAGACCAATTAAAAGGGTTCACAAGCCATGGACACAAATTAGCAGATCATGCCTTAGTTTTTATGGCAGACTATTTAAGACAGAATAAAGAATGGAGACAAGATTTCTTTGAAATTAATAAGAAAATAATTATACCTCTTTACGATGTTCCACATCTCATCAAGGGATTACGCAATAATCTCCTCACCAAAGATATGTATTATAAAGACGATGAAGATAAAGGCAAAATCAAAAAAGTGAAGTGGGAATATTTCCAATTGCTGTATGAGGCTGACAAATCATATGGAGAGTTAAGATGCCTTCAGAAATTAACAGAGGAGCATGTTAATCCTGATAAAATTAAGAAGATGCGAGTGAAGGCAGCAACGCAACTGTTCAGTCACAGTGTGGCTGTAGCCACTGAGCATTTAACAGCCAGAGGGAATTTGCCACAGGAGTGCAGGGATATAATTCCAATTGTATTAATAATTGACAAATTATTTGACAGTTTAAATGGTAACACTTTTCATGTACCCAACGGAAAACTACATAAAGGGCCTGTTAAAAGGATGCTGGGCAAAGGACATACGTGTGTGGCTGGGTTCTTACCAAATGTTTGA
Protein
MPRRCILKCAPEPGVRFHVFPSLESKPERFKAWVQAIGGKLDTPEDYEYYRNQRICDRHFAKENKNRYNRVDALGVPSLHLIETDLGEREKNSLGESFASSVQETLNLDVIEPTTSNVLVEDMMTRNNSLSLRKYNMYQSQIKQLRKRVNTFQKREDNFRTRLANAEKITDESAFKRIAKNMTLPAKIFTNMQFRTTSKKPSGRRFLLEEKVLSLTLYKRSPKCYSMLSRYFSLPAPKTLKRLLSEIKLGPGINTIVMEKLKKTVSELTMEDRLCTLIFDEMSITPQLHYDAAKDQLKGFTSHGHKLADHALVFMADYLRQNKEWRQDFFEINKKIIIPLYDVPHLIKGLRNNLLTKDMYYKDDEDKGKIKKVKWEYFQLLYEADKSYGELRCLQKLTEEHVNPDKIKKMRVKAATQLFSHSVAVATEHLTARGNLPQECRDIIPIVLIIDKLFDSLNGNTFHVPNGKLHKGPVKRMLGKGHTCVAGFLPNV
Summary
Uniprot
A0A3S2NSI9
A0A3S2KX83
A0A437BCW2
A0A3S2LM91
A0A3S2LWA3
A0A437ASS2
+ More
J9LY75 X1XB84 J9M4P5 J9M897 J9KX16 X1X0R9 A0A1Y1LY36 J9L7T1 A0A2A4JY71 J9KBW7 A0A2A4JZ58 A0A1Y1JZB1 A0A437AU19 J9KGM8 X1XNR4 J9LCP7 A0A1Y1NI49 J9L862 J9KCZ1 A0A437BL44 A0A1Y1KLF8 J9KI00 A0A3S2TA47 A0A2J7QSC0 J9L6Z3 A0A3S2LEZ0 X1XTB8
J9LY75 X1XB84 J9M4P5 J9M897 J9KX16 X1X0R9 A0A1Y1LY36 J9L7T1 A0A2A4JY71 J9KBW7 A0A2A4JZ58 A0A1Y1JZB1 A0A437AU19 J9KGM8 X1XNR4 J9LCP7 A0A1Y1NI49 J9L862 J9KCZ1 A0A437BL44 A0A1Y1KLF8 J9KI00 A0A3S2TA47 A0A2J7QSC0 J9L6Z3 A0A3S2LEZ0 X1XTB8
Pubmed
EMBL
RSAL01000257
RVE43416.1
RSAL01002057
RVE40596.1
RSAL01000087
RVE48224.1
+ More
RSAL01003176 RVE40327.1 RSAL01000161 RVE45426.1 RSAL01000699 RVE41218.1 ABLF02012658 ABLF02011758 ABLF02011759 ABLF02008684 ABLF02008686 ABLF02008687 ABLF02019592 ABLF02023911 ABLF02028653 ABLF02048521 ABLF02061836 ABLF02064667 GEZM01045715 JAV77678.1 ABLF02017696 NWSH01000363 PCG76967.1 ABLF02006763 PCG76968.1 GEZM01101736 JAV52346.1 RSAL01000475 RVE41592.1 ABLF02012220 ABLF02012222 ABLF02053218 ABLF02061299 ABLF02009194 GEZM01001796 JAV97521.1 ABLF02003317 ABLF02010389 RSAL01000037 RVE51172.1 GEZM01085096 JAV60236.1 ABLF02055473 ABLF02056006 RSAL01005638 RVE39985.1 NEVH01011876 PNF31489.1 ABLF02009647 ABLF02009778 ABLF02009779 ABLF02025341 ABLF02042390 RSAL01000165 RVE45277.1 ABLF02063349
RSAL01003176 RVE40327.1 RSAL01000161 RVE45426.1 RSAL01000699 RVE41218.1 ABLF02012658 ABLF02011758 ABLF02011759 ABLF02008684 ABLF02008686 ABLF02008687 ABLF02019592 ABLF02023911 ABLF02028653 ABLF02048521 ABLF02061836 ABLF02064667 GEZM01045715 JAV77678.1 ABLF02017696 NWSH01000363 PCG76967.1 ABLF02006763 PCG76968.1 GEZM01101736 JAV52346.1 RSAL01000475 RVE41592.1 ABLF02012220 ABLF02012222 ABLF02053218 ABLF02061299 ABLF02009194 GEZM01001796 JAV97521.1 ABLF02003317 ABLF02010389 RSAL01000037 RVE51172.1 GEZM01085096 JAV60236.1 ABLF02055473 ABLF02056006 RSAL01005638 RVE39985.1 NEVH01011876 PNF31489.1 ABLF02009647 ABLF02009778 ABLF02009779 ABLF02025341 ABLF02042390 RSAL01000165 RVE45277.1 ABLF02063349
Proteomes
Pfam
Interpro
IPR006816
ELMO_dom
+ More
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR021109 Peptidase_aspartic_dom_sf
IPR008737 Peptidase_asp_put
IPR006612 THAP_Znf
IPR005312 DUF1759
IPR021896 Transposase_37
IPR038441 THAP_Znf_sf
IPR005162 Retrotrans_gag_dom
IPR025398 DUF4371
IPR036691 Endo/exonu/phosph_ase_sf
IPR008906 HATC_C_dom
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR021109 Peptidase_aspartic_dom_sf
IPR008737 Peptidase_asp_put
IPR006612 THAP_Znf
IPR005312 DUF1759
IPR021896 Transposase_37
IPR038441 THAP_Znf_sf
IPR005162 Retrotrans_gag_dom
IPR025398 DUF4371
IPR036691 Endo/exonu/phosph_ase_sf
IPR008906 HATC_C_dom
Gene 3D
ProteinModelPortal
A0A3S2NSI9
A0A3S2KX83
A0A437BCW2
A0A3S2LM91
A0A3S2LWA3
A0A437ASS2
+ More
J9LY75 X1XB84 J9M4P5 J9M897 J9KX16 X1X0R9 A0A1Y1LY36 J9L7T1 A0A2A4JY71 J9KBW7 A0A2A4JZ58 A0A1Y1JZB1 A0A437AU19 J9KGM8 X1XNR4 J9LCP7 A0A1Y1NI49 J9L862 J9KCZ1 A0A437BL44 A0A1Y1KLF8 J9KI00 A0A3S2TA47 A0A2J7QSC0 J9L6Z3 A0A3S2LEZ0 X1XTB8
J9LY75 X1XB84 J9M4P5 J9M897 J9KX16 X1X0R9 A0A1Y1LY36 J9L7T1 A0A2A4JY71 J9KBW7 A0A2A4JZ58 A0A1Y1JZB1 A0A437AU19 J9KGM8 X1XNR4 J9LCP7 A0A1Y1NI49 J9L862 J9KCZ1 A0A437BL44 A0A1Y1KLF8 J9KI00 A0A3S2TA47 A0A2J7QSC0 J9L6Z3 A0A3S2LEZ0 X1XTB8
Ontologies
KEGG
GO
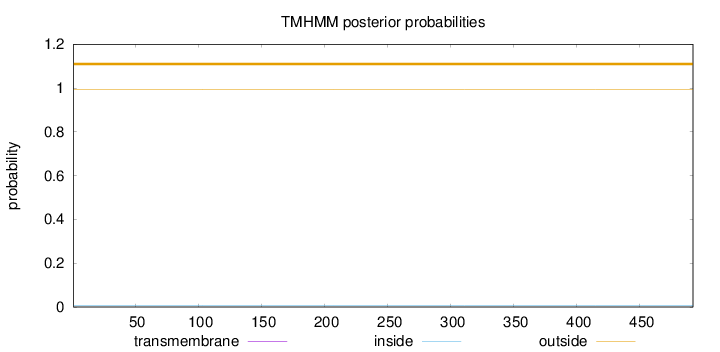
Topology
Length:
492
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00122
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00649
outside
1 - 492
Population Genetic Test Statistics
Pi
322.825661
Theta
184.835727
Tajima's D
2.231617
CLR
0.292354
CSRT
0.916454177291135
Interpretation
Uncertain
