Gene
KWMTBOMO08572
Pre Gene Modal
BGIBMGA007980
Annotation
FMRFamide_receptor_[Bombyx_mori]
Full name
FMRFamide receptor
Location in the cell
PlasmaMembrane Reliability : 4.87
Sequence
CDS
ATGAACGGCACCGAGGGGGAGGGGTGCGACTCCGACGTGGGCGTGCAGCCCGCCGACAAGCTGTTCCGCTTCGTGGTGCACGGCGTGCTGCTCAACGCGATCGGCGCTGCCGGCCTACTCGGCAACGCGCTGTCCGTCGTCGTGCTCTCGCGGCCACAGATGCGCTCCTCCATCAACTGTCTGCTGGTGGGGCTCGCCGCCTGCGACACCGTGCTCATCCTCACCTCCGTGCTGCTCTTTGGCCTCACCGCCGTTTACCCGTACACGGGCCGCCTGCGCTACTACTACTACCACGTCTGCCCCCACATCACGCCCTACGCCTACCCCATAGCCAACGCGGCGCAAACCATGTCCGTCTATCTAACGCTGATCGTGACGGTCGAGCGGTGGGTCGCCGTCTGCCATCCGTTCCGCGCCAAGTCGCTGTGCACGTCTTCGCGGGCCCGCTGGTACGTGCTGGGCACGGCCGCCTTCGCGCTCGCCTACAACGCTCCCAAGTTCCTCGAGGCGGAGGTGGTGACGCGCAGCGTGGACGGCGAGCCAGTGTACTGCGTGACGGCTGACCTGCACTTCCGCACCGAGACCTACATCGTGGTGTACATCCACTCCCTGTACATGATCGTGATGTACATCGTGCCGTTCTCTGCGCTGGCGGCGCTGAACGCGTGCATCGTGCGGCAGGTGCGGCGGGCGCAGGCCGAGCGGGCGCGCCTGTCGCGGGTGCAGCGCCGCGAGCTGGGCCTCGCCACCATGCTGCTCGTGGTCGTGCTCGTATTCTTCCTGTGCAACCTGCTGCCGCTCGTCACCAACTCGTTCGAAGTCTTCCTCGGGGACCAGCTAGAAAACTTAGATCCTCTCGTAAAGACGAGCAATCTTCTGGTGACAATCAACAGTAGCGTAAACTTTGTGATTTATGTGATATTTGGGGAGAAATTCAAGCGCGTGTTCCTGAAGATGTTCTGCGCGGGCGGATGGCGGCGGCGGACGCGCGACTCGCCGGAGCAGACGCGCGACGACTCGTTCGCGTCTTGCGGCGAGCGCGTGTCGCTGCGTCTAGTGCGGAACGGCACGCTGCGTCGCTCGGAGCTGCGCGCGCCCCCCCGGGGCCGCAGCCGCCGCGCCCCCTCCCCCTCCGTGTACTACCCCACCCCCCTCACTGACGTCACCAGCGCGCTCTCCGTCTCTGAGCCGCCGCCTGCCGCCATGCGATGGAACGGACACACGCGGTCGCACTTCTAA
Protein
MNGTEGEGCDSDVGVQPADKLFRFVVHGVLLNAIGAAGLLGNALSVVVLSRPQMRSSINCLLVGLAACDTVLILTSVLLFGLTAVYPYTGRLRYYYYHVCPHITPYAYPIANAAQTMSVYLTLIVTVERWVAVCHPFRAKSLCTSSRARWYVLGTAAFALAYNAPKFLEAEVVTRSVDGEPVYCVTADLHFRTETYIVVYIHSLYMIVMYIVPFSALAALNACIVRQVRRAQAERARLSRVQRRELGLATMLLVVVLVFFLCNLLPLVTNSFEVFLGDQLENLDPLVKTSNLLVTINSSVNFVIYVIFGEKFKRVFLKMFCAGGWRRRTRDSPEQTRDDSFASCGERVSLRLVRNGTLRRSELRAPPRGRSRRAPSPSVYYPTPLTDVTSALSVSEPPPAAMRWNGHTRSHF
Summary
Description
A receptor for the FMRFamide peptides. Reacts with high affinity to FMRFamide and intrinsic FMRFamide-related peptides.
Similarity
Belongs to the G-protein coupled receptor 1 family.
Keywords
Cell membrane
Complete proteome
G-protein coupled receptor
Glycoprotein
Membrane
Receptor
Reference proteome
Transducer
Transmembrane
Transmembrane helix
Feature
chain FMRFamide receptor
Uniprot
H9JEN3
Q1MW82
A0A2H1WR64
A0A2W1BE79
A0A0S1YDJ3
A0A194PVY8
+ More
A0A0N0PF39 A0A1J1IA62 A0A2J7RM31 A0A1B0D359 A0A067RIB2 A0A1B0CRX1 D6WCQ8 A0A182V7A9 A0A182PKD3 A0A182U6E5 A0A182KM77 A0A182I0I1 A0A182XBP5 A0A182Q005 Q7PYB7 A0A182YJX1 A0A182N4X7 A0A182VQV3 A0A182RBX4 E0VLX5 A0A182MKG8 Q1EHB5 A0A182H745 A0A084VRS3 A0A1Y1NB30 Q1DGJ7 Q17FI7 A0A182JK10 A0A1S4F4M7 U3U7P6 A0A182F6Y2 B0WKQ9 W5JQB2 A0A1B6M289 A0A1B6GSL8 A0A1B6HDC3 A0A154PN34 B4HTI3 B3M7F9 F4WSQ5 A0A2P8Y9A7 M9PBK3 Q9VZW5 B4QP04 A0A026VZK6 A0A1B0B700 A0A1A9YIC2 B4PCX0 Q29E43 B3NF13 X1WJ26 A0A1B0GFN0 A0A151ISG2 T1HBY9 A0A3B0JAT3 A0A0M5J5H2 F8J1K6 A0A310STP5 A0A151XDB0 E2AA00 A0A1W4VPK2 B4MN22 A0A0L7R555 A0A1I8M9G1 A0A0C9Q832 B4LE43 A0A2H8TIE0 A0A195AZH9 A0A1I8NYX2 A0A158NH31 A0A195D1G1 A0A195F5X6 B4IYI8 A0A0L0BPV9 A0A2S2R6Y2 B4KWL8 A0A088AK37 W8BCZ6 A0A2R7VSW2 A0A0A1X0K9 A0A0K8UUN5 A0A034VMD1 A0A0K2UX35 K7IRE2 A0A232F2W2 A0A3R7MSF5 E2BSD9 A0A3L8DN93 A0A336L365
A0A0N0PF39 A0A1J1IA62 A0A2J7RM31 A0A1B0D359 A0A067RIB2 A0A1B0CRX1 D6WCQ8 A0A182V7A9 A0A182PKD3 A0A182U6E5 A0A182KM77 A0A182I0I1 A0A182XBP5 A0A182Q005 Q7PYB7 A0A182YJX1 A0A182N4X7 A0A182VQV3 A0A182RBX4 E0VLX5 A0A182MKG8 Q1EHB5 A0A182H745 A0A084VRS3 A0A1Y1NB30 Q1DGJ7 Q17FI7 A0A182JK10 A0A1S4F4M7 U3U7P6 A0A182F6Y2 B0WKQ9 W5JQB2 A0A1B6M289 A0A1B6GSL8 A0A1B6HDC3 A0A154PN34 B4HTI3 B3M7F9 F4WSQ5 A0A2P8Y9A7 M9PBK3 Q9VZW5 B4QP04 A0A026VZK6 A0A1B0B700 A0A1A9YIC2 B4PCX0 Q29E43 B3NF13 X1WJ26 A0A1B0GFN0 A0A151ISG2 T1HBY9 A0A3B0JAT3 A0A0M5J5H2 F8J1K6 A0A310STP5 A0A151XDB0 E2AA00 A0A1W4VPK2 B4MN22 A0A0L7R555 A0A1I8M9G1 A0A0C9Q832 B4LE43 A0A2H8TIE0 A0A195AZH9 A0A1I8NYX2 A0A158NH31 A0A195D1G1 A0A195F5X6 B4IYI8 A0A0L0BPV9 A0A2S2R6Y2 B4KWL8 A0A088AK37 W8BCZ6 A0A2R7VSW2 A0A0A1X0K9 A0A0K8UUN5 A0A034VMD1 A0A0K2UX35 K7IRE2 A0A232F2W2 A0A3R7MSF5 E2BSD9 A0A3L8DN93 A0A336L365
Pubmed
19121390
16707581
28756777
26354079
24845553
18054377
+ More
18025266 18362917 18316733 20068045 19820115 21843505 23604020 20966253 12364791 14747013 17210077 25244985 20566863 26483478 24438588 28004739 17510324 30735632 23932938 20920257 23761445 17994087 18057021 21719571 29403074 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12218185 12438685 22936249 24508170 17550304 15632085 20798317 25315136 21347285 26108605 24495485 25830018 25348373 20075255 28648823 30249741
18025266 18362917 18316733 20068045 19820115 21843505 23604020 20966253 12364791 14747013 17210077 25244985 20566863 26483478 24438588 28004739 17510324 30735632 23932938 20920257 23761445 17994087 18057021 21719571 29403074 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12218185 12438685 22936249 24508170 17550304 15632085 20798317 25315136 21347285 26108605 24495485 25830018 25348373 20075255 28648823 30249741
EMBL
BABH01003378
AB253536
BAE93472.1
ODYU01010412
SOQ55497.1
KZ150358
+ More
PZC71216.1 KT031037 ALM88335.1 KQ459596 KPI95295.1 KQ459717 KPJ20327.1 CVRI01000047 CRK97175.1 CRK97179.1 NEVH01002577 PNF41890.1 AJVK01010888 KK852460 KDR23532.1 AJWK01025419 AJWK01025420 BK006097 KQ971317 DAA64478.1 EEZ99319.1 APCN01002079 AXCN02000187 AAAB01008987 EAA01756.2 DS235283 EEB14381.1 AXCM01003217 AY299455 AAQ73620.1 JXUM01115592 KQ565895 KXJ70560.1 ATLV01015774 KE525036 KFB40667.1 GEZM01010762 GEZM01010761 GEZM01010760 GEZM01010759 JAV93845.1 CH901475 EAT32276.2 CH477270 EAT45357.2 MH810178 QBC65443.1 AB817323 BAO01090.1 DS231975 EDS30001.1 ADMH02000641 ETN65478.1 GEBQ01009926 JAT30051.1 GECZ01004348 JAS65421.1 GECU01035023 JAS72683.1 KQ434998 KZC13291.1 CH480817 EDW50254.1 CH902618 EDV39857.1 KPU78249.1 GL888327 EGI62765.1 PYGN01000784 PSN40838.1 AE014296 AGB94042.1 AHN57950.1 AF351129 BK000442 AAF47700.1 AAL83921.1 DAA00378.1 CM000363 CM002912 EDX08999.1 KMY97215.1 KK107566 EZA48896.1 JXJN01009322 CM000159 EDW93874.1 CH379070 EAL30220.1 CH954178 EDV50355.1 ABLF02039963 ABLF02039969 ABLF02039979 CCAG010013759 KQ981091 KYN09544.1 ACPB03008224 OUUW01000002 SPP77112.1 CP012525 ALC43006.1 EU785049 ACI90286.1 KQ760357 OAD60755.1 KQ982294 KYQ58366.1 GL437990 EFN69729.1 CH963847 EDW73578.1 KQ414654 KOC65978.1 GBYB01010343 JAG80110.1 CH940647 EDW70086.1 GFXV01001966 MBW13771.1 KQ976698 KYM77447.1 ADTU01015499 KQ976973 KYN06686.1 KQ981768 KYN35873.1 CH916366 EDV97661.1 JRES01001546 KNC22120.1 GGMS01016257 MBY85460.1 CH933809 EDW19647.1 KRG06723.1 GAMC01015549 JAB91006.1 KK854028 PTY09770.1 GBXI01009861 JAD04431.1 GDHF01021895 GDHF01006817 JAI30419.1 JAI45497.1 GAKP01014456 JAC44496.1 HACA01025076 CDW42437.1 NNAY01001120 OXU25071.1 QCYY01000505 ROT84744.1 GL450203 EFN81391.1 QOIP01000006 RLU21914.1 UFQS01001431 UFQT01001431 SSX10853.1 SSX30533.1
PZC71216.1 KT031037 ALM88335.1 KQ459596 KPI95295.1 KQ459717 KPJ20327.1 CVRI01000047 CRK97175.1 CRK97179.1 NEVH01002577 PNF41890.1 AJVK01010888 KK852460 KDR23532.1 AJWK01025419 AJWK01025420 BK006097 KQ971317 DAA64478.1 EEZ99319.1 APCN01002079 AXCN02000187 AAAB01008987 EAA01756.2 DS235283 EEB14381.1 AXCM01003217 AY299455 AAQ73620.1 JXUM01115592 KQ565895 KXJ70560.1 ATLV01015774 KE525036 KFB40667.1 GEZM01010762 GEZM01010761 GEZM01010760 GEZM01010759 JAV93845.1 CH901475 EAT32276.2 CH477270 EAT45357.2 MH810178 QBC65443.1 AB817323 BAO01090.1 DS231975 EDS30001.1 ADMH02000641 ETN65478.1 GEBQ01009926 JAT30051.1 GECZ01004348 JAS65421.1 GECU01035023 JAS72683.1 KQ434998 KZC13291.1 CH480817 EDW50254.1 CH902618 EDV39857.1 KPU78249.1 GL888327 EGI62765.1 PYGN01000784 PSN40838.1 AE014296 AGB94042.1 AHN57950.1 AF351129 BK000442 AAF47700.1 AAL83921.1 DAA00378.1 CM000363 CM002912 EDX08999.1 KMY97215.1 KK107566 EZA48896.1 JXJN01009322 CM000159 EDW93874.1 CH379070 EAL30220.1 CH954178 EDV50355.1 ABLF02039963 ABLF02039969 ABLF02039979 CCAG010013759 KQ981091 KYN09544.1 ACPB03008224 OUUW01000002 SPP77112.1 CP012525 ALC43006.1 EU785049 ACI90286.1 KQ760357 OAD60755.1 KQ982294 KYQ58366.1 GL437990 EFN69729.1 CH963847 EDW73578.1 KQ414654 KOC65978.1 GBYB01010343 JAG80110.1 CH940647 EDW70086.1 GFXV01001966 MBW13771.1 KQ976698 KYM77447.1 ADTU01015499 KQ976973 KYN06686.1 KQ981768 KYN35873.1 CH916366 EDV97661.1 JRES01001546 KNC22120.1 GGMS01016257 MBY85460.1 CH933809 EDW19647.1 KRG06723.1 GAMC01015549 JAB91006.1 KK854028 PTY09770.1 GBXI01009861 JAD04431.1 GDHF01021895 GDHF01006817 JAI30419.1 JAI45497.1 GAKP01014456 JAC44496.1 HACA01025076 CDW42437.1 NNAY01001120 OXU25071.1 QCYY01000505 ROT84744.1 GL450203 EFN81391.1 QOIP01000006 RLU21914.1 UFQS01001431 UFQT01001431 SSX10853.1 SSX30533.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000183832
UP000235965
UP000092462
+ More
UP000027135 UP000092461 UP000007266 UP000075903 UP000075885 UP000075902 UP000075882 UP000075840 UP000076407 UP000075886 UP000007062 UP000076408 UP000075884 UP000075920 UP000075900 UP000009046 UP000075883 UP000069940 UP000249989 UP000030765 UP000008820 UP000075880 UP000069272 UP000002320 UP000000673 UP000076502 UP000001292 UP000007801 UP000007755 UP000245037 UP000000803 UP000000304 UP000053097 UP000092460 UP000092443 UP000002282 UP000001819 UP000008711 UP000007819 UP000092444 UP000078492 UP000015103 UP000268350 UP000092553 UP000075809 UP000000311 UP000192221 UP000007798 UP000053825 UP000095301 UP000008792 UP000078540 UP000095300 UP000005205 UP000078542 UP000078541 UP000001070 UP000037069 UP000009192 UP000005203 UP000002358 UP000215335 UP000283509 UP000008237 UP000279307
UP000027135 UP000092461 UP000007266 UP000075903 UP000075885 UP000075902 UP000075882 UP000075840 UP000076407 UP000075886 UP000007062 UP000076408 UP000075884 UP000075920 UP000075900 UP000009046 UP000075883 UP000069940 UP000249989 UP000030765 UP000008820 UP000075880 UP000069272 UP000002320 UP000000673 UP000076502 UP000001292 UP000007801 UP000007755 UP000245037 UP000000803 UP000000304 UP000053097 UP000092460 UP000092443 UP000002282 UP000001819 UP000008711 UP000007819 UP000092444 UP000078492 UP000015103 UP000268350 UP000092553 UP000075809 UP000000311 UP000192221 UP000007798 UP000053825 UP000095301 UP000008792 UP000078540 UP000095300 UP000005205 UP000078542 UP000078541 UP000001070 UP000037069 UP000009192 UP000005203 UP000002358 UP000215335 UP000283509 UP000008237 UP000279307
Interpro
ProteinModelPortal
H9JEN3
Q1MW82
A0A2H1WR64
A0A2W1BE79
A0A0S1YDJ3
A0A194PVY8
+ More
A0A0N0PF39 A0A1J1IA62 A0A2J7RM31 A0A1B0D359 A0A067RIB2 A0A1B0CRX1 D6WCQ8 A0A182V7A9 A0A182PKD3 A0A182U6E5 A0A182KM77 A0A182I0I1 A0A182XBP5 A0A182Q005 Q7PYB7 A0A182YJX1 A0A182N4X7 A0A182VQV3 A0A182RBX4 E0VLX5 A0A182MKG8 Q1EHB5 A0A182H745 A0A084VRS3 A0A1Y1NB30 Q1DGJ7 Q17FI7 A0A182JK10 A0A1S4F4M7 U3U7P6 A0A182F6Y2 B0WKQ9 W5JQB2 A0A1B6M289 A0A1B6GSL8 A0A1B6HDC3 A0A154PN34 B4HTI3 B3M7F9 F4WSQ5 A0A2P8Y9A7 M9PBK3 Q9VZW5 B4QP04 A0A026VZK6 A0A1B0B700 A0A1A9YIC2 B4PCX0 Q29E43 B3NF13 X1WJ26 A0A1B0GFN0 A0A151ISG2 T1HBY9 A0A3B0JAT3 A0A0M5J5H2 F8J1K6 A0A310STP5 A0A151XDB0 E2AA00 A0A1W4VPK2 B4MN22 A0A0L7R555 A0A1I8M9G1 A0A0C9Q832 B4LE43 A0A2H8TIE0 A0A195AZH9 A0A1I8NYX2 A0A158NH31 A0A195D1G1 A0A195F5X6 B4IYI8 A0A0L0BPV9 A0A2S2R6Y2 B4KWL8 A0A088AK37 W8BCZ6 A0A2R7VSW2 A0A0A1X0K9 A0A0K8UUN5 A0A034VMD1 A0A0K2UX35 K7IRE2 A0A232F2W2 A0A3R7MSF5 E2BSD9 A0A3L8DN93 A0A336L365
A0A0N0PF39 A0A1J1IA62 A0A2J7RM31 A0A1B0D359 A0A067RIB2 A0A1B0CRX1 D6WCQ8 A0A182V7A9 A0A182PKD3 A0A182U6E5 A0A182KM77 A0A182I0I1 A0A182XBP5 A0A182Q005 Q7PYB7 A0A182YJX1 A0A182N4X7 A0A182VQV3 A0A182RBX4 E0VLX5 A0A182MKG8 Q1EHB5 A0A182H745 A0A084VRS3 A0A1Y1NB30 Q1DGJ7 Q17FI7 A0A182JK10 A0A1S4F4M7 U3U7P6 A0A182F6Y2 B0WKQ9 W5JQB2 A0A1B6M289 A0A1B6GSL8 A0A1B6HDC3 A0A154PN34 B4HTI3 B3M7F9 F4WSQ5 A0A2P8Y9A7 M9PBK3 Q9VZW5 B4QP04 A0A026VZK6 A0A1B0B700 A0A1A9YIC2 B4PCX0 Q29E43 B3NF13 X1WJ26 A0A1B0GFN0 A0A151ISG2 T1HBY9 A0A3B0JAT3 A0A0M5J5H2 F8J1K6 A0A310STP5 A0A151XDB0 E2AA00 A0A1W4VPK2 B4MN22 A0A0L7R555 A0A1I8M9G1 A0A0C9Q832 B4LE43 A0A2H8TIE0 A0A195AZH9 A0A1I8NYX2 A0A158NH31 A0A195D1G1 A0A195F5X6 B4IYI8 A0A0L0BPV9 A0A2S2R6Y2 B4KWL8 A0A088AK37 W8BCZ6 A0A2R7VSW2 A0A0A1X0K9 A0A0K8UUN5 A0A034VMD1 A0A0K2UX35 K7IRE2 A0A232F2W2 A0A3R7MSF5 E2BSD9 A0A3L8DN93 A0A336L365
PDB
4ZJC
E-value=8.37173e-07,
Score=127
Ontologies
GO
Topology
Subcellular location
Cell membrane
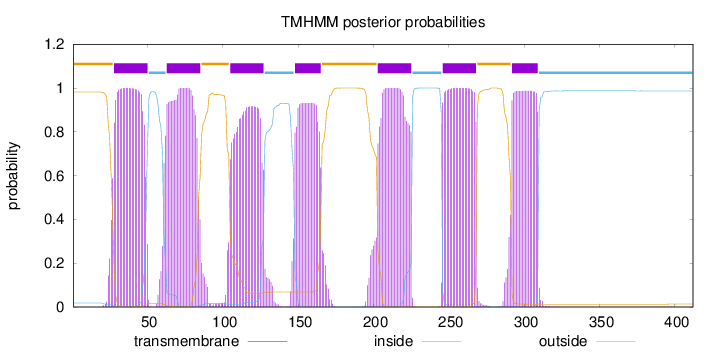
Length:
412
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
150.11517
Exp number, first 60 AAs:
23.35322
Total prob of N-in:
0.01788
POSSIBLE N-term signal
sequence
outside
1 - 27
TMhelix
28 - 50
inside
51 - 62
TMhelix
63 - 85
outside
86 - 104
TMhelix
105 - 127
inside
128 - 147
TMhelix
148 - 165
outside
166 - 202
TMhelix
203 - 225
inside
226 - 245
TMhelix
246 - 268
outside
269 - 291
TMhelix
292 - 309
inside
310 - 412
Population Genetic Test Statistics
Pi
67.246843
Theta
53.624961
Tajima's D
-0.634137
CLR
17.576446
CSRT
0.212739363031848
Interpretation
Uncertain
