Pre Gene Modal
BGIBMGA013806
Annotation
PREDICTED:_uncharacterized_protein_LOC105841662_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.314
Sequence
CDS
ATGTACCGCACACCCACGACTTCGGGCGCCGCTAAAAACCCTACCACTCCCAGCCCTGCTACCACAACCGCAGTGGCGCAGGGTGCTGCAAAGACGTTTATCAAAAGACCGGACACCAAAAGAAGATCACTGGATAATATGGACGTTTATAAAAAGCAGCAACAAAACTACGCTGCACTGCCTAGTGATCAGCTCCTCCAGGAGATAATGAAGACTTTTGAGGACATACCCTCGAAAGTTCAAGAGTACCCTCAGATGAGGAGGGATATCAAACAGTTCCTGGAGGTGCAGTCAGATAAGGGTATACGAGCAATCTTCGAGCTCAGTCGGCGCATCCAAAGTCTCGAAGCAGAGGCCCCGAAGACAAGGACACTTGCAGAAACCTCGTCGTCTTATGTCCAAGATATCATACTTGGGAGGCTGGGAGAGGTAGAAACGACACTGACTGAGCGACTGAACGAGATGGAGAGAAGTCTGAAAAGGGACATAATACAACTGAACGACGAAAATGACGGCCGGGGCTCCGACATGATCGACAAGATCATCGACAGGATCGACGAGACCAGAGCTGCCATGGACCAAACGACCGAGGCGGTCGAGAGTCGAACGCACGCTCTCGTCAAGGATCTGGTCGGTGGTCTTGCCGATGATGTCGTTGCAGCAAAACAAGAACTTACGGAGGTCGCAAAGGGACTGGCATGCGATATAAACCTAGCTCCAGTCTCTAAGTCGTACGCCAGCGTCGCAGCAGCCAAACCCGTCCACCGTCCTGCACTTCATTCGATGGCAGTCACTCTCGAAGGTAGTCAGGAGACTGCTGACGAGGTGCTCGCCAGGGTGAAGAGGGCAGTCGACGCCAGAGGCACTGGACTTAAAATTAACAAAGTACGCAAGGCAAAAAACCAAACTATTATATTGGGTTGCGACACCGAGGAGGAAAGAAAGAAAGTCCAAGAGATAATAGAGAGTCGTGGGGATGGACTGGTAGTAGCGCCCATGAAGAACAAGAGCCCTCTCGTTGTCCTGAAGGATGTTTTCGTGGAAAGTGAAGACGACGATATGATTAAGGCACTCCACGTCCAAAACAAGAACATCTTCATGGGCCTACCGGAAGAAGACAGGCAAATGGAGATAAAATTTAAAAAACGTACCAGAAACCCAAAGACCGCGCACATCATCTTGCAAGTGAAGCCGAAAGTATGGCAGAGGATGACGGAAGCTGGAGCCCTCTATCTGGACCTTCAAAGGGTCAGAGTCGAGGACCAGTCTCCGCTCATCCAATGCACGAGGTGCCTTGCATTTGGACATGGTCGTAAATTTTGCACCGAGAGTGTGGACAGATGCAGTCACTGTGGAGGACCGCATCTGCGCGAAAAATGCGCAGACTTTATCGCAGGGACCGAACCGCGGTGCTGCAACTGCTCACACTCTGGGTTGCGGAAGGCTGACCACAACGCTTTTAGCGCCGAATGCCCGGTACGAAAAAAGTGGGACTACTTGGCTCGCGCAAATACCACATATGCATGA
Protein
MYRTPTTSGAAKNPTTPSPATTTAVAQGAAKTFIKRPDTKRRSLDNMDVYKKQQQNYAALPSDQLLQEIMKTFEDIPSKVQEYPQMRRDIKQFLEVQSDKGIRAIFELSRRIQSLEAEAPKTRTLAETSSSYVQDIILGRLGEVETTLTERLNEMERSLKRDIIQLNDENDGRGSDMIDKIIDRIDETRAAMDQTTEAVESRTHALVKDLVGGLADDVVAAKQELTEVAKGLACDINLAPVSKSYASVAAAKPVHRPALHSMAVTLEGSQETADEVLARVKRAVDARGTGLKINKVRKAKNQTIILGCDTEEERKKVQEIIESRGDGLVVAPMKNKSPLVVLKDVFVESEDDDMIKALHVQNKNIFMGLPEEDRQMEIKFKKRTRNPKTAHIILQVKPKVWQRMTEAGALYLDLQRVRVEDQSPLIQCTRCLAFGHGRKFCTESVDRCSHCGGPHLREKCADFIAGTEPRCCNCSHSGLRKADHNAFSAECPVRKKWDYLARANTTYA
Summary
Uniprot
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
Ontologies
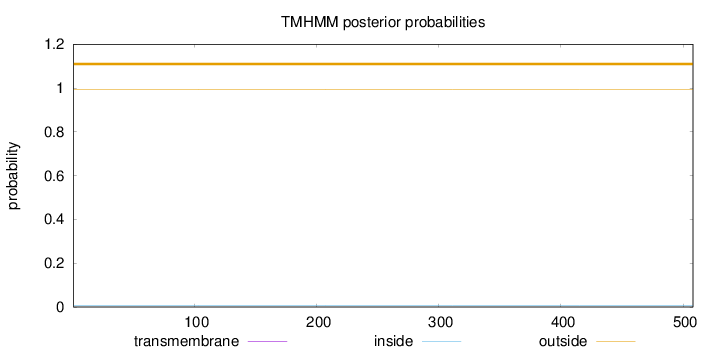
Topology
Length:
508
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00135
Exp number, first 60 AAs:
0.00135
Total prob of N-in:
0.00574
outside
1 - 508
Population Genetic Test Statistics
Pi
74.986418
Theta
116.744602
Tajima's D
0.25893
CLR
0
CSRT
0.446927653617319
Interpretation
Uncertain
