Gene
KWMTBOMO08570
Annotation
unnamed_protein_product_[Tribolium_castaneum]
Location in the cell
Cytoplasmic Reliability : 1.489 Mitochondrial Reliability : 1.806
Sequence
CDS
ATGGGCATGGCACAAATTCAAAGCATAGCTGATGGCCTCTTCCCGGAACACATAGATTTCGAACGGAAAACGAAACCGTCACAGGAGTTCCCAGAGGTGACCGAGGAGGAGATCATCCAACTAAAAGCTCGCATAAAGCTCAGGAAAGCCCCAGGGCCCGACCGGATTCCGCCGGAGATAGTGACTCTAGCAGTAGCAGTAATTCCAGACAGGATAAGGCTTGTGTTGCAGAAGGTGCTGGAGTCTGGCGTCTTCCCCAACAGCTGGAAAACAGCCAGACTAGTCCTGCTAAGAAAACCAGGCAAACCCGCTGAAGATCCTGCGGGGCACAGGCCGCTATGTCTACTGGACTGCTACGGAAAGCTGCTGGAGTCGCTGATCCTAGGCAGAATAGAAAAAACAACTACCAACAAGCTACTTGAGATAGCGGAAGACGCGAGACGAGGAACCCACCGCACCAGAAGAATCTGCGCAGTAGTGGCCCTCGACGTCCGTAATGCTTTCAATTCTGCATCGTGGGAGCTCATCCTGGAGGAAATGGCTGCAATAGGTATGCCTCAATACATCTATAATATCATAAACAGCTACCTCCAGGACAGATGCATCATACTGACAGACGCTGAGGGAGGGGTTGTAGTGAAAAAGGTTACCGGTGGTGTTCCGCAGGGGTCGATACTGGGCCCCACGCTGTGGAACATACTGTATGATGGGGTACTGCGCCTCGAACTGGGGGATGGGGCACAATTAATAGGCTTCGCGGACGATTTAGCTCTGGTGGTGTCAGCAAAGAAGGAAGCAGAACTAATGGCAATAACGAATGTAGCCCTACAAAAGATCAGCGCTTGGATGAAGCAAAAGCGGCTACACTTGGCCCCAGAAAAGACCGAGGCAATTGTGCTAAGTGGGCGAAGAAAGCTTAGCGAGATTTGCTTTACCCTCCAGGGCGTCAGGGTGGTTCCAACCGACAAACTGAAATACCTCGGGTTTTGGTTTGACAAAAGTCTGAAGTTCGGCAAACACATTGAAGAAACAACTGGTAAAGCCGAAAGACTGACGACCGCCCTTGGCCAAATCATGCCCAGGAAAGGAGGGCCGAGGTCGAGCAAGCGCAGACTATTGGCGTCCGCAATACAATCTGTGATGTTATATGGAGCACCGGTGTGGGCAAAGGTAATGAATGTAGCGAAGTACAGGAACATGTATAGCAAAGTGCCAACGACAGCTGGCAATACGCATCTGTGGAGCTTACCGTACCATATCGCTAGACGCCGTCTTGGTGATAGCGGGCCTGATACCGATAGAACGGCTAGCAAAGGAACCGGCAACGCTACTACCACGAGGGGACAAGAACGAGAGAAGAGGAAAGAGCACATACAATAA
Protein
MGMAQIQSIADGLFPEHIDFERKTKPSQEFPEVTEEEIIQLKARIKLRKAPGPDRIPPEIVTLAVAVIPDRIRLVLQKVLESGVFPNSWKTARLVLLRKPGKPAEDPAGHRPLCLLDCYGKLLESLILGRIEKTTTNKLLEIAEDARRGTHRTRRICAVVALDVRNAFNSASWELILEEMAAIGMPQYIYNIINSYLQDRCIILTDAEGGVVVKKVTGGVPQGSILGPTLWNILYDGVLRLELGDGAQLIGFADDLALVVSAKKEAELMAITNVALQKISAWMKQKRLHLAPEKTEAIVLSGRRKLSEICFTLQGVRVVPTDKLKYLGFWFDKSLKFGKHIEETTGKAERLTTALGQIMPRKGGPRSSKRRLLASAIQSVMLYGAPVWAKVMNVAKYRNMYSKVPTTAGNTHLWSLPYHIARRRLGDSGPDTDRTASKGTGNATTTRGQEREKRKEHIQ
Summary
Uniprot
A0A1B6KDF3
F7IYV5
D7GXX9
A0A1W7R6C5
A0A139W8G2
F7IYU8
+ More
V5I881 A0A139WAH4 A0A139WN93 A0A139WNC9 A0A142LX45 A0A139W940 T1DCM0 T1DG59 A0A139W9B3 T1DG89 A0A139W939 A0A139W9J0 A0A1S4EMG7 D6WC19 A0A0K8TR60 J9KUM6 W8AMZ9 A0A2M4CL98 J9KJG8 V5GWW9 A0A2S2N8I2 A0A2H8TR36 A0A1W7R6I9 J9L6Z9 A0A2M4CS98 A0A023EW96 J9KT61 A0A2M4AGH2 F7IYV3 N6TYN9 A0A0E3W2C5 A0A2M4AHG6 A0A2M4ADW9 F7IYV2 J9L0J0 W8ADE4 W8ANK7 A0A034WR48 V5I8F3 J9KV04 U4TRW5 A0A2M4BDI3 A0A2M4BC01 A0A2M4CKQ7 A0A2M4BBV9 J9LVU7 A0A139W9A6 A0A2S2NW56 A0A142LX37 X1WQ45 A0A0K8S5I5 A0A2M4BC28 J9LBG0 A0A2S2P488 V9GZD9 K7JP45 A0A2S2PRL8 X1X6F1 Q868S4 Q868S0 A0A2M4BCE1 N6UIP2 A0A2M4CJY9 A0A1S3DCR6 Q868T0 A0A224XIZ2 J9KNN8 A0A2M4CSA9 J9LQQ1 A0A2M4CSI6 A0A139W9I2 A0A2M3Z1V8 A0A2H8TIE5 J9LBB9 X1WZ64 A0A139W8J5 A0A2M3Z1C1 F7IYV7 A0A2M4BB78 A0A2M4BB89 J9KM84 Q9N9Z1 J9LM37 A0A2M4AK35 A0A142LX43 A0A2M4BC18 J9KNG8 J9JZR8 D7EIB0 A0A2M4CJ59 A0A2M4BC24 A0A2M4CKD9 V5G2Q2 A0A2M4CJ33
V5I881 A0A139WAH4 A0A139WN93 A0A139WNC9 A0A142LX45 A0A139W940 T1DCM0 T1DG59 A0A139W9B3 T1DG89 A0A139W939 A0A139W9J0 A0A1S4EMG7 D6WC19 A0A0K8TR60 J9KUM6 W8AMZ9 A0A2M4CL98 J9KJG8 V5GWW9 A0A2S2N8I2 A0A2H8TR36 A0A1W7R6I9 J9L6Z9 A0A2M4CS98 A0A023EW96 J9KT61 A0A2M4AGH2 F7IYV3 N6TYN9 A0A0E3W2C5 A0A2M4AHG6 A0A2M4ADW9 F7IYV2 J9L0J0 W8ADE4 W8ANK7 A0A034WR48 V5I8F3 J9KV04 U4TRW5 A0A2M4BDI3 A0A2M4BC01 A0A2M4CKQ7 A0A2M4BBV9 J9LVU7 A0A139W9A6 A0A2S2NW56 A0A142LX37 X1WQ45 A0A0K8S5I5 A0A2M4BC28 J9LBG0 A0A2S2P488 V9GZD9 K7JP45 A0A2S2PRL8 X1X6F1 Q868S4 Q868S0 A0A2M4BCE1 N6UIP2 A0A2M4CJY9 A0A1S3DCR6 Q868T0 A0A224XIZ2 J9KNN8 A0A2M4CSA9 J9LQQ1 A0A2M4CSI6 A0A139W9I2 A0A2M3Z1V8 A0A2H8TIE5 J9LBB9 X1WZ64 A0A139W8J5 A0A2M3Z1C1 F7IYV7 A0A2M4BB78 A0A2M4BB89 J9KM84 Q9N9Z1 J9LM37 A0A2M4AK35 A0A142LX43 A0A2M4BC18 J9KNG8 J9JZR8 D7EIB0 A0A2M4CJ59 A0A2M4BC24 A0A2M4CKD9 V5G2Q2 A0A2M4CJ33
Pubmed
EMBL
GEBQ01030504
JAT09473.1
AB593325
BAK38646.1
KQ972160
EFA13619.2
+ More
GEHC01000964 JAV46681.1 KQ973343 KXZ75566.1 AB593321 BAK38639.1 GALX01005145 JAB63321.1 KQ971388 KYB24913.1 KQ971310 KYB29499.1 KYB29500.1 KU543683 AMS38371.1 KQ972691 KXZ75806.1 GALA01001777 JAA93075.1 GALA01000302 JAA94550.1 KQ972544 KXZ75869.1 GALA01001776 JAA93076.1 KXZ75807.1 KQ972226 KYB24577.1 KQ971309 EEZ99146.1 GDAI01000741 JAI16862.1 ABLF02041557 GAMC01019173 JAB87382.1 GGFL01001916 MBW66094.1 ABLF02041886 GALX01002229 JAB66237.1 GGMR01000779 MBY13398.1 GFXV01003903 MBW15708.1 GEHC01000915 JAV46730.1 ABLF02021100 GGFL01004044 MBW68222.1 GAPW01000192 JAC13406.1 ABLF02009073 ABLF02030702 ABLF02042963 GGFK01006563 MBW39884.1 AB593324 BAK38644.1 APGK01005465 KB736249 ENN83233.1 HACL01000281 CFW94575.1 GGFK01006866 MBW40187.1 GGFK01005660 MBW38981.1 AB593323 BAK38643.1 ABLF02027248 GAMC01020519 GAMC01020515 JAB86040.1 GAMC01020517 JAB86038.1 GAKP01002292 JAC56660.1 GALX01004785 JAB63681.1 ABLF02004674 ABLF02004675 ABLF02004676 ABLF02011637 KB630188 ERL83462.1 GGFJ01001727 MBW50868.1 GGFJ01001370 MBW50511.1 GGFL01001583 MBW65761.1 GGFJ01001369 MBW50510.1 ABLF02034925 KXZ75870.1 GGMR01008673 MBY21292.1 KU543679 AMS38363.1 ABLF02004920 ABLF02004921 ABLF02015191 ABLF02016615 ABLF02024214 GBRD01017378 JAG48449.1 GGFJ01001439 MBW50580.1 ABLF02021645 GGMR01011682 MBY24301.1 M93691 AAA29365.1 AAZX01001144 GGMR01019493 MBY32112.1 ABLF02016340 AB090815 BAC57906.1 AB090817 BAC57910.1 GGFJ01001541 MBW50682.1 APGK01033685 KB740885 ENN78512.1 GGFL01001456 MBW65634.1 AB090812 BAC57900.1 GFTR01008427 JAW07999.1 ABLF02023257 GGFL01004046 MBW68224.1 ABLF02030663 ABLF02041312 GGFL01004047 MBW68225.1 KQ972255 KYB24568.1 GGFM01001733 MBW22484.1 GFXV01002090 MBW13895.1 ABLF02020562 ABLF02011183 ABLF02041884 KQ973256 KXZ75595.1 GGFM01001554 MBW22305.1 AB593327 BAK38648.1 GGFJ01001164 MBW50305.1 GGFJ01001165 MBW50306.1 ABLF02018942 ABLF02041885 AJ278684 CAB99192.1 ABLF02014650 GGFK01007771 MBW41092.1 KU543682 AMS38369.1 GGFJ01001453 MBW50594.1 ABLF02005203 ABLF02041316 ABLF02009709 ABLF02009711 AB593322 BAK38641.1 GGFL01001189 MBW65367.1 GGFJ01001454 MBW50595.1 GGFL01001190 MBW65368.1 GALX01004171 JAB64295.1 GGFL01001192 MBW65370.1
GEHC01000964 JAV46681.1 KQ973343 KXZ75566.1 AB593321 BAK38639.1 GALX01005145 JAB63321.1 KQ971388 KYB24913.1 KQ971310 KYB29499.1 KYB29500.1 KU543683 AMS38371.1 KQ972691 KXZ75806.1 GALA01001777 JAA93075.1 GALA01000302 JAA94550.1 KQ972544 KXZ75869.1 GALA01001776 JAA93076.1 KXZ75807.1 KQ972226 KYB24577.1 KQ971309 EEZ99146.1 GDAI01000741 JAI16862.1 ABLF02041557 GAMC01019173 JAB87382.1 GGFL01001916 MBW66094.1 ABLF02041886 GALX01002229 JAB66237.1 GGMR01000779 MBY13398.1 GFXV01003903 MBW15708.1 GEHC01000915 JAV46730.1 ABLF02021100 GGFL01004044 MBW68222.1 GAPW01000192 JAC13406.1 ABLF02009073 ABLF02030702 ABLF02042963 GGFK01006563 MBW39884.1 AB593324 BAK38644.1 APGK01005465 KB736249 ENN83233.1 HACL01000281 CFW94575.1 GGFK01006866 MBW40187.1 GGFK01005660 MBW38981.1 AB593323 BAK38643.1 ABLF02027248 GAMC01020519 GAMC01020515 JAB86040.1 GAMC01020517 JAB86038.1 GAKP01002292 JAC56660.1 GALX01004785 JAB63681.1 ABLF02004674 ABLF02004675 ABLF02004676 ABLF02011637 KB630188 ERL83462.1 GGFJ01001727 MBW50868.1 GGFJ01001370 MBW50511.1 GGFL01001583 MBW65761.1 GGFJ01001369 MBW50510.1 ABLF02034925 KXZ75870.1 GGMR01008673 MBY21292.1 KU543679 AMS38363.1 ABLF02004920 ABLF02004921 ABLF02015191 ABLF02016615 ABLF02024214 GBRD01017378 JAG48449.1 GGFJ01001439 MBW50580.1 ABLF02021645 GGMR01011682 MBY24301.1 M93691 AAA29365.1 AAZX01001144 GGMR01019493 MBY32112.1 ABLF02016340 AB090815 BAC57906.1 AB090817 BAC57910.1 GGFJ01001541 MBW50682.1 APGK01033685 KB740885 ENN78512.1 GGFL01001456 MBW65634.1 AB090812 BAC57900.1 GFTR01008427 JAW07999.1 ABLF02023257 GGFL01004046 MBW68224.1 ABLF02030663 ABLF02041312 GGFL01004047 MBW68225.1 KQ972255 KYB24568.1 GGFM01001733 MBW22484.1 GFXV01002090 MBW13895.1 ABLF02020562 ABLF02011183 ABLF02041884 KQ973256 KXZ75595.1 GGFM01001554 MBW22305.1 AB593327 BAK38648.1 GGFJ01001164 MBW50305.1 GGFJ01001165 MBW50306.1 ABLF02018942 ABLF02041885 AJ278684 CAB99192.1 ABLF02014650 GGFK01007771 MBW41092.1 KU543682 AMS38369.1 GGFJ01001453 MBW50594.1 ABLF02005203 ABLF02041316 ABLF02009709 ABLF02009711 AB593322 BAK38641.1 GGFL01001189 MBW65367.1 GGFJ01001454 MBW50595.1 GGFL01001190 MBW65368.1 GALX01004171 JAB64295.1 GGFL01001192 MBW65370.1
Proteomes
PRIDE
Pfam
Interpro
Gene 3D
ProteinModelPortal
A0A1B6KDF3
F7IYV5
D7GXX9
A0A1W7R6C5
A0A139W8G2
F7IYU8
+ More
V5I881 A0A139WAH4 A0A139WN93 A0A139WNC9 A0A142LX45 A0A139W940 T1DCM0 T1DG59 A0A139W9B3 T1DG89 A0A139W939 A0A139W9J0 A0A1S4EMG7 D6WC19 A0A0K8TR60 J9KUM6 W8AMZ9 A0A2M4CL98 J9KJG8 V5GWW9 A0A2S2N8I2 A0A2H8TR36 A0A1W7R6I9 J9L6Z9 A0A2M4CS98 A0A023EW96 J9KT61 A0A2M4AGH2 F7IYV3 N6TYN9 A0A0E3W2C5 A0A2M4AHG6 A0A2M4ADW9 F7IYV2 J9L0J0 W8ADE4 W8ANK7 A0A034WR48 V5I8F3 J9KV04 U4TRW5 A0A2M4BDI3 A0A2M4BC01 A0A2M4CKQ7 A0A2M4BBV9 J9LVU7 A0A139W9A6 A0A2S2NW56 A0A142LX37 X1WQ45 A0A0K8S5I5 A0A2M4BC28 J9LBG0 A0A2S2P488 V9GZD9 K7JP45 A0A2S2PRL8 X1X6F1 Q868S4 Q868S0 A0A2M4BCE1 N6UIP2 A0A2M4CJY9 A0A1S3DCR6 Q868T0 A0A224XIZ2 J9KNN8 A0A2M4CSA9 J9LQQ1 A0A2M4CSI6 A0A139W9I2 A0A2M3Z1V8 A0A2H8TIE5 J9LBB9 X1WZ64 A0A139W8J5 A0A2M3Z1C1 F7IYV7 A0A2M4BB78 A0A2M4BB89 J9KM84 Q9N9Z1 J9LM37 A0A2M4AK35 A0A142LX43 A0A2M4BC18 J9KNG8 J9JZR8 D7EIB0 A0A2M4CJ59 A0A2M4BC24 A0A2M4CKD9 V5G2Q2 A0A2M4CJ33
V5I881 A0A139WAH4 A0A139WN93 A0A139WNC9 A0A142LX45 A0A139W940 T1DCM0 T1DG59 A0A139W9B3 T1DG89 A0A139W939 A0A139W9J0 A0A1S4EMG7 D6WC19 A0A0K8TR60 J9KUM6 W8AMZ9 A0A2M4CL98 J9KJG8 V5GWW9 A0A2S2N8I2 A0A2H8TR36 A0A1W7R6I9 J9L6Z9 A0A2M4CS98 A0A023EW96 J9KT61 A0A2M4AGH2 F7IYV3 N6TYN9 A0A0E3W2C5 A0A2M4AHG6 A0A2M4ADW9 F7IYV2 J9L0J0 W8ADE4 W8ANK7 A0A034WR48 V5I8F3 J9KV04 U4TRW5 A0A2M4BDI3 A0A2M4BC01 A0A2M4CKQ7 A0A2M4BBV9 J9LVU7 A0A139W9A6 A0A2S2NW56 A0A142LX37 X1WQ45 A0A0K8S5I5 A0A2M4BC28 J9LBG0 A0A2S2P488 V9GZD9 K7JP45 A0A2S2PRL8 X1X6F1 Q868S4 Q868S0 A0A2M4BCE1 N6UIP2 A0A2M4CJY9 A0A1S3DCR6 Q868T0 A0A224XIZ2 J9KNN8 A0A2M4CSA9 J9LQQ1 A0A2M4CSI6 A0A139W9I2 A0A2M3Z1V8 A0A2H8TIE5 J9LBB9 X1WZ64 A0A139W8J5 A0A2M3Z1C1 F7IYV7 A0A2M4BB78 A0A2M4BB89 J9KM84 Q9N9Z1 J9LM37 A0A2M4AK35 A0A142LX43 A0A2M4BC18 J9KNG8 J9JZR8 D7EIB0 A0A2M4CJ59 A0A2M4BC24 A0A2M4CKD9 V5G2Q2 A0A2M4CJ33
Ontologies
GO
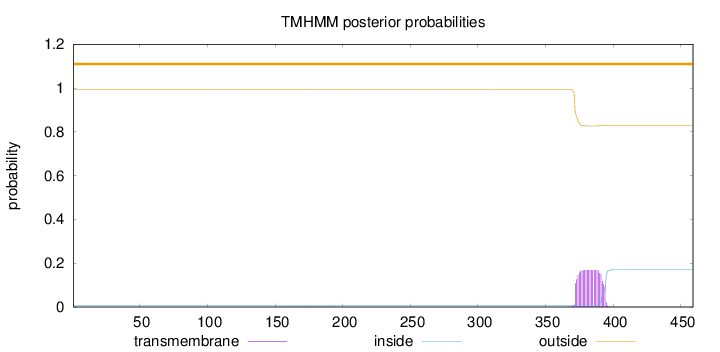
Topology
Length:
459
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.56771
Exp number, first 60 AAs:
0.00021
Total prob of N-in:
0.00745
outside
1 - 459
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
130.658719
CSRT
0
Interpretation
Uncertain
