Gene
KWMTBOMO08567
Pre Gene Modal
BGIBMGA009307
Annotation
PREDICTED:_thioredoxin-like_protein_4A_[Bombyx_mori]
Full name
Thioredoxin-like protein
Location in the cell
Cytoplasmic Reliability : 3.2
Sequence
CDS
ATGAGTTATATGCTAGGACATCTACACAATGGTTGGCAAGTAGATCAGGCCATTCTATCAGAAGAAGACCGTGTAGTGGTTATAAGGTTTGGACACGACTGGGACCCAACATGTATGAAAATGGATGAAGTATTATACAGCATTGCGGAAAAAGTAAAAAATTTTGCTGTCATATATTTAGTTGACATTACAGAAGTTCCAGATTTTAATAAAATGTATGAATTATACGATCCATGCACAGTGATGTTCTTCTTCCGCAACAAACACATAATGATTGACTTGGGGACTGGCAACAACAATAAAATCAACTGGCCTCTCGAAGACAAGCAGGAGATGATTGATATTATTGAAACTGTGTACAGAGGGGCAAGAAAAGGACGAGGTCTTGTTGTCTCACCGAAAGATTACTCTACCAAATACAGATATTAG
Protein
MSYMLGHLHNGWQVDQAILSEEDRVVVIRFGHDWDPTCMKMDEVLYSIAEKVKNFAVIYLVDITEVPDFNKMYELYDPCTVMFFFRNKHIMIDLGTGNNNKINWPLEDKQEMIDIIETVYRGARKGRGLVVSPKDYSTKYRY
Summary
Description
Plays role in pre-mRNA splicing.
Similarity
Belongs to the DIM1 family.
Uniprot
A0A2A4J717
A0A194RB12
A0A194Q725
H9JIF7
S4PTT3
A0A212FFU0
+ More
M4JSD8 Q1HQS6 A0A182G8J6 A0A1Q3G2K7 A0A0L7LFT5 E0VF07 A0A2J7PWU8 A0A067RGH2 A0A2P8XHQ8 D2A3I2 A0A443QMQ0 A0A182MV62 A0A182RMD6 A0A182QJ52 B0WIR3 A0A1B6FFU1 A0A1B6K1I0 A0A401S022 A0A401PJ37 K4FT74 N6UPC7 A0A1J1IMT0 A0A1A9XJM2 A0A1A9VW13 A0A1B0BCW6 A0A1A9ZCT5 A0A1I8NBC0 A0A0L0CHP3 A0A0K8TNP0 A0A084VXZ9 A0A182JCD9 T1E8Y7 U5EMR1 A0A182Y3F2 A0A182PCQ8 A0A2M3ZCW6 A0A2M4AV46 A0A2M4C2P4 A0A182UZ11 A0A182MZ06 W5JAR6 A0A182KDL8 A0A182W6N7 A0A182UHD5 A0A182XL73 A0A182L0W4 Q7Q496 A0A182ICF4 A0A3B3D624 A0A3B3HCV1 A0A437CU22 A0A3P9K893 A0A452TE24 A0A1Z5L9G2 A0A1W4WAM0 B4LT76 A0A0J9QVM5 B4KES0 A0A3B0K2P8 Q29LJ5 B4GPV4 A0A034WVD8 B4NYK0 A0A0M3QTE6 A0A1I8PA76 B3MU37 B4MZM2 B4I387 B4JCM2 B3N3R8 A0A0A1XKY1 A0A0K8VZC2 Q8SZ87 A0A1B0GMU9 A0A0K8SY97 A0A1S4EIW1 A0A0C9R976 A0A2S2N4H7 G1MTF6 A0A151NFX5 A0A3L8SMA5 A0A093S5S7 A0A091FRU6 A0A091W0E6 A0A3M0KSL0 K7FC77 A0A1V4KEF4 A0A091P4G4 U3KFU6 A0A0Q3PPD1 A0A0A0A1Q0 A0A1U7S8P1 A0A087QXZ4 A0A452GXA8
M4JSD8 Q1HQS6 A0A182G8J6 A0A1Q3G2K7 A0A0L7LFT5 E0VF07 A0A2J7PWU8 A0A067RGH2 A0A2P8XHQ8 D2A3I2 A0A443QMQ0 A0A182MV62 A0A182RMD6 A0A182QJ52 B0WIR3 A0A1B6FFU1 A0A1B6K1I0 A0A401S022 A0A401PJ37 K4FT74 N6UPC7 A0A1J1IMT0 A0A1A9XJM2 A0A1A9VW13 A0A1B0BCW6 A0A1A9ZCT5 A0A1I8NBC0 A0A0L0CHP3 A0A0K8TNP0 A0A084VXZ9 A0A182JCD9 T1E8Y7 U5EMR1 A0A182Y3F2 A0A182PCQ8 A0A2M3ZCW6 A0A2M4AV46 A0A2M4C2P4 A0A182UZ11 A0A182MZ06 W5JAR6 A0A182KDL8 A0A182W6N7 A0A182UHD5 A0A182XL73 A0A182L0W4 Q7Q496 A0A182ICF4 A0A3B3D624 A0A3B3HCV1 A0A437CU22 A0A3P9K893 A0A452TE24 A0A1Z5L9G2 A0A1W4WAM0 B4LT76 A0A0J9QVM5 B4KES0 A0A3B0K2P8 Q29LJ5 B4GPV4 A0A034WVD8 B4NYK0 A0A0M3QTE6 A0A1I8PA76 B3MU37 B4MZM2 B4I387 B4JCM2 B3N3R8 A0A0A1XKY1 A0A0K8VZC2 Q8SZ87 A0A1B0GMU9 A0A0K8SY97 A0A1S4EIW1 A0A0C9R976 A0A2S2N4H7 G1MTF6 A0A151NFX5 A0A3L8SMA5 A0A093S5S7 A0A091FRU6 A0A091W0E6 A0A3M0KSL0 K7FC77 A0A1V4KEF4 A0A091P4G4 U3KFU6 A0A0Q3PPD1 A0A0A0A1Q0 A0A1U7S8P1 A0A087QXZ4 A0A452GXA8
Pubmed
26354079
19121390
23622113
22118469
23473738
17204158
+ More
17510324 26483478 26227816 20566863 24845553 29403074 18362917 19820115 30297745 23056606 23537049 25315136 26108605 26369729 24438588 25244985 20920257 23761445 20966253 12364791 14747013 17210077 29451363 17554307 24813606 28528879 17994087 22936249 15632085 25348373 17550304 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26823975 20838655 22293439 30282656 17381049 28562605
17510324 26483478 26227816 20566863 24845553 29403074 18362917 19820115 30297745 23056606 23537049 25315136 26108605 26369729 24438588 25244985 20920257 23761445 20966253 12364791 14747013 17210077 29451363 17554307 24813606 28528879 17994087 22936249 15632085 25348373 17550304 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26823975 20838655 22293439 30282656 17381049 28562605
EMBL
NWSH01002949
PCG67240.1
KQ460436
KPJ14689.1
KQ459337
KPJ01347.1
+ More
BABH01027282 GAIX01011488 JAA81072.1 AGBW02008766 OWR52598.1 JX867717 ODYU01012954 AGC94855.1 SOQ59510.1 DQ440368 CH477389 ABF18401.1 EAT42010.1 JXUM01047822 KQ561534 KXJ78243.1 GFDL01000990 JAV34055.1 JTDY01001364 KOB74071.1 DS235100 EEB11981.1 NEVH01020867 PNF20806.1 KK852480 KDR22966.1 PYGN01002093 PSN31526.1 KQ971338 EFA02875.1 NCKU01005647 RWS04314.1 AXCM01010852 AXCN02000789 DS231951 EDS28640.1 GECZ01020693 JAS49076.1 GECU01002411 JAT05296.1 BEZZ01000038 GCC23761.1 BFAA01000596 GCB73153.1 JX052717 AFK10945.1 APGK01011543 KB738503 KB632168 ENN82581.1 ERL89396.1 CVRI01000054 CRL00852.1 JXJN01012155 JRES01000385 KNC31737.1 GDAI01001596 JAI16007.1 ATLV01018234 KE525226 KFB42843.1 AXCP01000428 GAMD01002343 JAA99247.1 GANO01000908 JAB58963.1 GGFM01005646 MBW26397.1 GGFK01011320 MBW44641.1 GGFJ01010441 MBW59582.1 ADMH02002027 GGFL01004237 ETN59879.1 MBW68415.1 AAAB01008964 EAA12234.1 APCN01000798 CM012447 RVE66452.1 GFJQ02002924 JAW04046.1 CH940649 EDW64918.1 CM002910 KMY88061.1 CH933807 EDW12970.1 OUUW01000004 SPP79886.1 CH379060 EAL34050.1 CH479187 EDW39626.1 GAKP01000650 JAC58302.1 CM000157 EDW87633.1 CP012523 ALC38749.1 CH902624 EDV33366.1 CH963920 EDW77807.1 CH480820 EDW54232.1 CH916368 EDW04186.1 CH954177 EDV57727.1 GBXI01003094 JAD11198.1 GDHF01016577 GDHF01015459 GDHF01008076 JAI35737.1 JAI36855.1 JAI44238.1 AE014134 AY071048 AAF51017.2 AAL48670.1 AJVK01004176 GBRD01007627 GDHC01000833 JAG58194.1 JAQ17796.1 GBYB01003336 JAG73103.1 GGMT01000314 MBY12000.1 AKHW03003201 KYO35425.1 QUSF01000013 RLW04356.1 KL670090 KFW78269.1 KL447416 KFO73240.1 KL411540 KFR09147.1 QRBI01000104 RMC16289.1 AGCU01158196 AGCU01158197 LSYS01003422 OPJ82785.1 KK844575 KFP86450.1 AGTO01021309 LMAW01001669 KQK82923.1 KL870344 KGL87897.1 KL225997 KFM06098.1
BABH01027282 GAIX01011488 JAA81072.1 AGBW02008766 OWR52598.1 JX867717 ODYU01012954 AGC94855.1 SOQ59510.1 DQ440368 CH477389 ABF18401.1 EAT42010.1 JXUM01047822 KQ561534 KXJ78243.1 GFDL01000990 JAV34055.1 JTDY01001364 KOB74071.1 DS235100 EEB11981.1 NEVH01020867 PNF20806.1 KK852480 KDR22966.1 PYGN01002093 PSN31526.1 KQ971338 EFA02875.1 NCKU01005647 RWS04314.1 AXCM01010852 AXCN02000789 DS231951 EDS28640.1 GECZ01020693 JAS49076.1 GECU01002411 JAT05296.1 BEZZ01000038 GCC23761.1 BFAA01000596 GCB73153.1 JX052717 AFK10945.1 APGK01011543 KB738503 KB632168 ENN82581.1 ERL89396.1 CVRI01000054 CRL00852.1 JXJN01012155 JRES01000385 KNC31737.1 GDAI01001596 JAI16007.1 ATLV01018234 KE525226 KFB42843.1 AXCP01000428 GAMD01002343 JAA99247.1 GANO01000908 JAB58963.1 GGFM01005646 MBW26397.1 GGFK01011320 MBW44641.1 GGFJ01010441 MBW59582.1 ADMH02002027 GGFL01004237 ETN59879.1 MBW68415.1 AAAB01008964 EAA12234.1 APCN01000798 CM012447 RVE66452.1 GFJQ02002924 JAW04046.1 CH940649 EDW64918.1 CM002910 KMY88061.1 CH933807 EDW12970.1 OUUW01000004 SPP79886.1 CH379060 EAL34050.1 CH479187 EDW39626.1 GAKP01000650 JAC58302.1 CM000157 EDW87633.1 CP012523 ALC38749.1 CH902624 EDV33366.1 CH963920 EDW77807.1 CH480820 EDW54232.1 CH916368 EDW04186.1 CH954177 EDV57727.1 GBXI01003094 JAD11198.1 GDHF01016577 GDHF01015459 GDHF01008076 JAI35737.1 JAI36855.1 JAI44238.1 AE014134 AY071048 AAF51017.2 AAL48670.1 AJVK01004176 GBRD01007627 GDHC01000833 JAG58194.1 JAQ17796.1 GBYB01003336 JAG73103.1 GGMT01000314 MBY12000.1 AKHW03003201 KYO35425.1 QUSF01000013 RLW04356.1 KL670090 KFW78269.1 KL447416 KFO73240.1 KL411540 KFR09147.1 QRBI01000104 RMC16289.1 AGCU01158196 AGCU01158197 LSYS01003422 OPJ82785.1 KK844575 KFP86450.1 AGTO01021309 LMAW01001669 KQK82923.1 KL870344 KGL87897.1 KL225997 KFM06098.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000005204
UP000007151
UP000008820
+ More
UP000069940 UP000249989 UP000037510 UP000009046 UP000235965 UP000027135 UP000245037 UP000007266 UP000285301 UP000075883 UP000075900 UP000075886 UP000002320 UP000287033 UP000288216 UP000019118 UP000030742 UP000183832 UP000092443 UP000078200 UP000092460 UP000092445 UP000095301 UP000037069 UP000030765 UP000075880 UP000076408 UP000075885 UP000075903 UP000075884 UP000000673 UP000075881 UP000075920 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000261560 UP000001038 UP000265180 UP000265200 UP000291021 UP000192221 UP000008792 UP000009192 UP000268350 UP000001819 UP000008744 UP000002282 UP000092553 UP000095300 UP000007801 UP000007798 UP000001292 UP000001070 UP000008711 UP000000803 UP000092462 UP000079169 UP000001645 UP000050525 UP000276834 UP000053258 UP000053760 UP000053283 UP000269221 UP000007267 UP000190648 UP000016665 UP000051836 UP000053858 UP000189705 UP000053286 UP000291020
UP000069940 UP000249989 UP000037510 UP000009046 UP000235965 UP000027135 UP000245037 UP000007266 UP000285301 UP000075883 UP000075900 UP000075886 UP000002320 UP000287033 UP000288216 UP000019118 UP000030742 UP000183832 UP000092443 UP000078200 UP000092460 UP000092445 UP000095301 UP000037069 UP000030765 UP000075880 UP000076408 UP000075885 UP000075903 UP000075884 UP000000673 UP000075881 UP000075920 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000261560 UP000001038 UP000265180 UP000265200 UP000291021 UP000192221 UP000008792 UP000009192 UP000268350 UP000001819 UP000008744 UP000002282 UP000092553 UP000095300 UP000007801 UP000007798 UP000001292 UP000001070 UP000008711 UP000000803 UP000092462 UP000079169 UP000001645 UP000050525 UP000276834 UP000053258 UP000053760 UP000053283 UP000269221 UP000007267 UP000190648 UP000016665 UP000051836 UP000053858 UP000189705 UP000053286 UP000291020
Pfam
PF02966 DIM1
SUPFAM
SSF52833
SSF52833
CDD
ProteinModelPortal
A0A2A4J717
A0A194RB12
A0A194Q725
H9JIF7
S4PTT3
A0A212FFU0
+ More
M4JSD8 Q1HQS6 A0A182G8J6 A0A1Q3G2K7 A0A0L7LFT5 E0VF07 A0A2J7PWU8 A0A067RGH2 A0A2P8XHQ8 D2A3I2 A0A443QMQ0 A0A182MV62 A0A182RMD6 A0A182QJ52 B0WIR3 A0A1B6FFU1 A0A1B6K1I0 A0A401S022 A0A401PJ37 K4FT74 N6UPC7 A0A1J1IMT0 A0A1A9XJM2 A0A1A9VW13 A0A1B0BCW6 A0A1A9ZCT5 A0A1I8NBC0 A0A0L0CHP3 A0A0K8TNP0 A0A084VXZ9 A0A182JCD9 T1E8Y7 U5EMR1 A0A182Y3F2 A0A182PCQ8 A0A2M3ZCW6 A0A2M4AV46 A0A2M4C2P4 A0A182UZ11 A0A182MZ06 W5JAR6 A0A182KDL8 A0A182W6N7 A0A182UHD5 A0A182XL73 A0A182L0W4 Q7Q496 A0A182ICF4 A0A3B3D624 A0A3B3HCV1 A0A437CU22 A0A3P9K893 A0A452TE24 A0A1Z5L9G2 A0A1W4WAM0 B4LT76 A0A0J9QVM5 B4KES0 A0A3B0K2P8 Q29LJ5 B4GPV4 A0A034WVD8 B4NYK0 A0A0M3QTE6 A0A1I8PA76 B3MU37 B4MZM2 B4I387 B4JCM2 B3N3R8 A0A0A1XKY1 A0A0K8VZC2 Q8SZ87 A0A1B0GMU9 A0A0K8SY97 A0A1S4EIW1 A0A0C9R976 A0A2S2N4H7 G1MTF6 A0A151NFX5 A0A3L8SMA5 A0A093S5S7 A0A091FRU6 A0A091W0E6 A0A3M0KSL0 K7FC77 A0A1V4KEF4 A0A091P4G4 U3KFU6 A0A0Q3PPD1 A0A0A0A1Q0 A0A1U7S8P1 A0A087QXZ4 A0A452GXA8
M4JSD8 Q1HQS6 A0A182G8J6 A0A1Q3G2K7 A0A0L7LFT5 E0VF07 A0A2J7PWU8 A0A067RGH2 A0A2P8XHQ8 D2A3I2 A0A443QMQ0 A0A182MV62 A0A182RMD6 A0A182QJ52 B0WIR3 A0A1B6FFU1 A0A1B6K1I0 A0A401S022 A0A401PJ37 K4FT74 N6UPC7 A0A1J1IMT0 A0A1A9XJM2 A0A1A9VW13 A0A1B0BCW6 A0A1A9ZCT5 A0A1I8NBC0 A0A0L0CHP3 A0A0K8TNP0 A0A084VXZ9 A0A182JCD9 T1E8Y7 U5EMR1 A0A182Y3F2 A0A182PCQ8 A0A2M3ZCW6 A0A2M4AV46 A0A2M4C2P4 A0A182UZ11 A0A182MZ06 W5JAR6 A0A182KDL8 A0A182W6N7 A0A182UHD5 A0A182XL73 A0A182L0W4 Q7Q496 A0A182ICF4 A0A3B3D624 A0A3B3HCV1 A0A437CU22 A0A3P9K893 A0A452TE24 A0A1Z5L9G2 A0A1W4WAM0 B4LT76 A0A0J9QVM5 B4KES0 A0A3B0K2P8 Q29LJ5 B4GPV4 A0A034WVD8 B4NYK0 A0A0M3QTE6 A0A1I8PA76 B3MU37 B4MZM2 B4I387 B4JCM2 B3N3R8 A0A0A1XKY1 A0A0K8VZC2 Q8SZ87 A0A1B0GMU9 A0A0K8SY97 A0A1S4EIW1 A0A0C9R976 A0A2S2N4H7 G1MTF6 A0A151NFX5 A0A3L8SMA5 A0A093S5S7 A0A091FRU6 A0A091W0E6 A0A3M0KSL0 K7FC77 A0A1V4KEF4 A0A091P4G4 U3KFU6 A0A0Q3PPD1 A0A0A0A1Q0 A0A1U7S8P1 A0A087QXZ4 A0A452GXA8
PDB
6QX9
E-value=3.17324e-77,
Score=727
Ontologies
GO
PANTHER
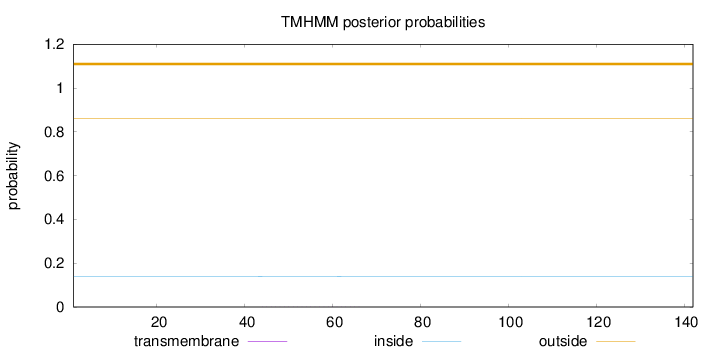
Topology
Subcellular location
Nucleus
Length:
142
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00986
Exp number, first 60 AAs:
0.00736
Total prob of N-in:
0.13883
outside
1 - 142
Population Genetic Test Statistics
Pi
272.44504
Theta
195.682663
Tajima's D
1.143642
CLR
0
CSRT
0.694215289235538
Interpretation
Uncertain
