Gene
KWMTBOMO08565
Pre Gene Modal
BGIBMGA009308
Annotation
PREDICTED:_protein_cereblon-like_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.24 Nuclear Reliability : 1.084
Sequence
CDS
ATGGATCTAGATGAGAACTCACTTGATAGCGAATCGCTGCAAAGAGTCTCTGCTAGTGAAGAAGTGGATGACGAGCAGACAACCTCAGAAGATGAGCAAGACTTTGACATCTCTTTAGCTGCCTCGCATTCCTACATGGGCGGAGGGCTCAGCTCGGTGCGAGGGCGCGTGGTGCTGGAGGCGGGCTGGGAGGGCCGCGTGCCGGTGGTGGCGCACCACGGCGCCGTGTTCCCGGGAGAGACCGTGCCCATGCTCGTGTCCAACGCGCACGACGCCGCCATCGTACAGAAAATCAAGGACGACAAACTCTTCGGCTTGCTGTGCCCCGACGGCGACAACGGCGCGGCGGTGTCGGGGTACGGCGCGCTGTGCGAGGTGTACGAGGCGGCGCTGGGCGGGGCCGCCGAGCCCGCCGCGCTCAGCTTCAAGGCGCGCGCCACGCACCGCTTCCGCTTCTGCCACATGCCCAAGTCTTCCGTGCCCATACACCTCTACGCCAGACTGCGGCTGGCGGACGTGCGCGTGCTGCCCGACGTGTGCGGCCCCCCGCCGCTGCGCGCCCCCGGCCTGGACTCGCTCAGGTCGCGGCGGGGGCTGCGCTACGTGGACGCGGCGCTCACGGCGTGGCCCGCGTTCGTCTACGACATGTTCGACTACGAGCGCATGCGGAACATCATCAAGGCGTACTTCGACACGCTGATGCTGGACGCGCTGCCGGAGGGCGCCGTGTCGCTGTCGTACTGGGTGGCGTCCAACCTGACGCTGGGCGCGCGCGACCGCCTGGCGCTGTTCGCGGTGGACGACGCGCTGCTGCGGCTGCACCTCGAGGTGCGCTTCATCACTCAGTTCTCCCGCGCCCAGAAGTGCGCGCTGTGCTGCGGGTCCTGCGGCGGGGAGGTGGCGCAGCGCGAGCACGTGTTCCCCATGTCCAGCCACGGCGTGCACTCCAACTACACCAACCCGGGCGGGTACGTGCACGGCGTGGTGACGGTGACGCGCGCGCGCGCCGCGCCCGGCGGCCCGCCCTCCGCCGACTACTCGTGGTTCCCCGGCTACAGCTGGACCGTCGCGCTGTGCCGAGACTGCCGCGCGCACGTGGGCTGGAGGTTCGACGCCATAAAGAGGAGCCTCCGCCCGCAGCAGTTCTACGGGCTCTGCAGGAACTACGTGCAGCCGCGACACCACGACCAGGACCTCGCCGACTACTGA
Protein
MDLDENSLDSESLQRVSASEEVDDEQTTSEDEQDFDISLAASHSYMGGGLSSVRGRVVLEAGWEGRVPVVAHHGAVFPGETVPMLVSNAHDAAIVQKIKDDKLFGLLCPDGDNGAAVSGYGALCEVYEAALGGAAEPAALSFKARATHRFRFCHMPKSSVPIHLYARLRLADVRVLPDVCGPPPLRAPGLDSLRSRRGLRYVDAALTAWPAFVYDMFDYERMRNIIKAYFDTLMLDALPEGAVSLSYWVASNLTLGARDRLALFAVDDALLRLHLEVRFITQFSRAQKCALCCGSCGGEVAQREHVFPMSSHGVHSNYTNPGGYVHGVVTVTRARAAPGGPPSADYSWFPGYSWTVALCRDCRAHVGWRFDAIKRSLRPQQFYGLCRNYVQPRHHDQDLADY
Summary
Uniprot
H9JIF8
A0A212FFT0
A0A437B5T3
A0A2A4K207
A0A2A4K2N0
A0A194RA26
+ More
A0A195CE96 F4WYQ7 A0A151JZ65 A0A158N947 A0A026W788 A0A3L8DJ29 E2AK45 A0A151X4N5 A0A182ITC4 A0A336K518 Q17B57 A0A195BVM7 A0A182HD31 A0A084VXM2 A0A2A3EFL3 A0A088A4J1 B0W9N6 A0A210QI98 A0A0N0BFL6 A0A1Q3G4B2 Q17B58 E2C926 W5JFG0 A0A182TGD6 A0A1Q3FA94 A0A182FCZ4 K1QI70 A0A182PJ24 A0A182RQE1 Q17B59 A0A1S4G7L3 Q7QI48 A0A182UW62 A0A182WRZ8 A0A182ICX1 A0A1L8DTY0 A0A182KVW8 A0A182JW43 A0A182QRU2 A0A182MDL1 C3Z2Q1 A0A182NN59 A0A232FDX4 A0A293M3Q0 A0A1L8DTN8 K7IW26 A0A1Z5LGI1 A0A182HD29 A0A182SR45 A0A2R5L7E3 A0A182WK39 T1JIH3 Q4RXV6 H3DAT4 V9KHU4 H2TVC3 G3N6P5 A0A3B5KIU4 A0A3Q4MZ43 A0A3P9P9B6 A0A3P8USH6 A0A131XY44 B7PWV7 A0A3B5MQG3 A0A3B3QYH1 W5MZ09 A0A1B6E425 A0A1W5A8W1 A0A154PMJ9 A7S6W7 A0A3P8TFF6 A0A3B3QXW8 A0A3Q1HNM0 A0A3Q1CAZ9 A0A0L8HQK9 R7TQ36 A0A3Q0TBH3 A0A3Q3QXI8 A0A0L8HQR6 A0A0S7GTZ3 A0A3B3X2F1 A0A3B5QUV4 I3KS87 A0A3Q2CF56 A0A3B4F8U9 A0A3Q2WK80 A0A3P9BQM6
A0A195CE96 F4WYQ7 A0A151JZ65 A0A158N947 A0A026W788 A0A3L8DJ29 E2AK45 A0A151X4N5 A0A182ITC4 A0A336K518 Q17B57 A0A195BVM7 A0A182HD31 A0A084VXM2 A0A2A3EFL3 A0A088A4J1 B0W9N6 A0A210QI98 A0A0N0BFL6 A0A1Q3G4B2 Q17B58 E2C926 W5JFG0 A0A182TGD6 A0A1Q3FA94 A0A182FCZ4 K1QI70 A0A182PJ24 A0A182RQE1 Q17B59 A0A1S4G7L3 Q7QI48 A0A182UW62 A0A182WRZ8 A0A182ICX1 A0A1L8DTY0 A0A182KVW8 A0A182JW43 A0A182QRU2 A0A182MDL1 C3Z2Q1 A0A182NN59 A0A232FDX4 A0A293M3Q0 A0A1L8DTN8 K7IW26 A0A1Z5LGI1 A0A182HD29 A0A182SR45 A0A2R5L7E3 A0A182WK39 T1JIH3 Q4RXV6 H3DAT4 V9KHU4 H2TVC3 G3N6P5 A0A3B5KIU4 A0A3Q4MZ43 A0A3P9P9B6 A0A3P8USH6 A0A131XY44 B7PWV7 A0A3B5MQG3 A0A3B3QYH1 W5MZ09 A0A1B6E425 A0A1W5A8W1 A0A154PMJ9 A7S6W7 A0A3P8TFF6 A0A3B3QXW8 A0A3Q1HNM0 A0A3Q1CAZ9 A0A0L8HQK9 R7TQ36 A0A3Q0TBH3 A0A3Q3QXI8 A0A0L8HQR6 A0A0S7GTZ3 A0A3B3X2F1 A0A3B5QUV4 I3KS87 A0A3Q2CF56 A0A3B4F8U9 A0A3Q2WK80 A0A3P9BQM6
Pubmed
EMBL
BABH01027277
BABH01027278
BABH01027279
AGBW02008766
OWR52595.1
RSAL01000154
+ More
RVE45669.1 NWSH01000219 PCG78277.1 PCG78276.1 KQ460436 KPJ14693.1 KQ977935 KYM98556.1 GL888459 EGI60668.1 KQ981440 KYN41667.1 ADTU01009091 ADTU01009092 ADTU01009093 ADTU01009094 ADTU01009095 KK107364 EZA51920.1 QOIP01000007 RLU20417.1 GL440191 EFN66188.1 KQ982543 KYQ55383.1 UFQS01000100 UFQT01000100 SSW99551.1 SSX19931.1 CH477325 EAT43511.1 KQ976405 KYM91851.1 JXUM01005144 KQ560187 KXJ83934.1 ATLV01018097 KE525212 KFB42716.1 KZ288266 PBC30274.1 DS231865 EDS40355.1 NEDP02003604 OWF48321.1 KQ435800 KOX73264.1 GFDL01000407 JAV34638.1 EAT43510.1 GL453768 EFN75562.1 ADMH02001370 ETN62811.1 GFDL01010617 JAV24428.1 JH817484 EKC30879.1 EAT43509.1 AAAB01008810 EAA04790.4 APCN01006058 GFDF01004339 JAV09745.1 AXCN02001952 AXCM01001616 GG666574 EEN53302.1 NNAY01000393 OXU28688.1 GFWV01010645 MAA35374.1 GFDF01004340 JAV09744.1 GFJQ02000712 JAW06258.1 JXUM01005139 KXJ83932.1 GGLE01001308 MBY05434.1 JH431701 CAAE01014979 CAG06776.1 JW865083 AFO97600.1 GEFM01003867 JAP71929.1 ABJB010035570 DS810469 EEC11079.1 AHAT01031660 GEDC01004633 JAS32665.1 KQ434948 KZC12440.1 DS471342 DS469590 EDO28630.1 EDO40533.1 KQ417510 KOF91521.1 AMQN01002386 KB309044 ELT95677.1 KOF91519.1 GBYX01449383 GBYX01449377 JAO32104.1 AERX01006711
RVE45669.1 NWSH01000219 PCG78277.1 PCG78276.1 KQ460436 KPJ14693.1 KQ977935 KYM98556.1 GL888459 EGI60668.1 KQ981440 KYN41667.1 ADTU01009091 ADTU01009092 ADTU01009093 ADTU01009094 ADTU01009095 KK107364 EZA51920.1 QOIP01000007 RLU20417.1 GL440191 EFN66188.1 KQ982543 KYQ55383.1 UFQS01000100 UFQT01000100 SSW99551.1 SSX19931.1 CH477325 EAT43511.1 KQ976405 KYM91851.1 JXUM01005144 KQ560187 KXJ83934.1 ATLV01018097 KE525212 KFB42716.1 KZ288266 PBC30274.1 DS231865 EDS40355.1 NEDP02003604 OWF48321.1 KQ435800 KOX73264.1 GFDL01000407 JAV34638.1 EAT43510.1 GL453768 EFN75562.1 ADMH02001370 ETN62811.1 GFDL01010617 JAV24428.1 JH817484 EKC30879.1 EAT43509.1 AAAB01008810 EAA04790.4 APCN01006058 GFDF01004339 JAV09745.1 AXCN02001952 AXCM01001616 GG666574 EEN53302.1 NNAY01000393 OXU28688.1 GFWV01010645 MAA35374.1 GFDF01004340 JAV09744.1 GFJQ02000712 JAW06258.1 JXUM01005139 KXJ83932.1 GGLE01001308 MBY05434.1 JH431701 CAAE01014979 CAG06776.1 JW865083 AFO97600.1 GEFM01003867 JAP71929.1 ABJB010035570 DS810469 EEC11079.1 AHAT01031660 GEDC01004633 JAS32665.1 KQ434948 KZC12440.1 DS471342 DS469590 EDO28630.1 EDO40533.1 KQ417510 KOF91521.1 AMQN01002386 KB309044 ELT95677.1 KOF91519.1 GBYX01449383 GBYX01449377 JAO32104.1 AERX01006711
Proteomes
UP000005204
UP000007151
UP000283053
UP000218220
UP000053240
UP000078542
+ More
UP000007755 UP000078541 UP000005205 UP000053097 UP000279307 UP000000311 UP000075809 UP000075880 UP000008820 UP000078540 UP000069940 UP000249989 UP000030765 UP000242457 UP000005203 UP000002320 UP000242188 UP000053105 UP000008237 UP000000673 UP000075902 UP000069272 UP000005408 UP000075885 UP000075900 UP000007062 UP000075903 UP000076407 UP000075840 UP000075882 UP000075881 UP000075886 UP000075883 UP000001554 UP000075884 UP000215335 UP000002358 UP000075901 UP000075920 UP000007303 UP000005226 UP000007635 UP000261580 UP000242638 UP000265120 UP000001555 UP000261380 UP000261540 UP000018468 UP000192224 UP000076502 UP000001593 UP000265080 UP000257200 UP000257160 UP000053454 UP000014760 UP000261340 UP000261600 UP000261480 UP000002852 UP000005207 UP000265020 UP000261460 UP000264840 UP000265160
UP000007755 UP000078541 UP000005205 UP000053097 UP000279307 UP000000311 UP000075809 UP000075880 UP000008820 UP000078540 UP000069940 UP000249989 UP000030765 UP000242457 UP000005203 UP000002320 UP000242188 UP000053105 UP000008237 UP000000673 UP000075902 UP000069272 UP000005408 UP000075885 UP000075900 UP000007062 UP000075903 UP000076407 UP000075840 UP000075882 UP000075881 UP000075886 UP000075883 UP000001554 UP000075884 UP000215335 UP000002358 UP000075901 UP000075920 UP000007303 UP000005226 UP000007635 UP000261580 UP000242638 UP000265120 UP000001555 UP000261380 UP000261540 UP000018468 UP000192224 UP000076502 UP000001593 UP000265080 UP000257200 UP000257160 UP000053454 UP000014760 UP000261340 UP000261600 UP000261480 UP000002852 UP000005207 UP000265020 UP000261460 UP000264840 UP000265160
PRIDE
Interpro
SUPFAM
SSF88697
SSF88697
ProteinModelPortal
H9JIF8
A0A212FFT0
A0A437B5T3
A0A2A4K207
A0A2A4K2N0
A0A194RA26
+ More
A0A195CE96 F4WYQ7 A0A151JZ65 A0A158N947 A0A026W788 A0A3L8DJ29 E2AK45 A0A151X4N5 A0A182ITC4 A0A336K518 Q17B57 A0A195BVM7 A0A182HD31 A0A084VXM2 A0A2A3EFL3 A0A088A4J1 B0W9N6 A0A210QI98 A0A0N0BFL6 A0A1Q3G4B2 Q17B58 E2C926 W5JFG0 A0A182TGD6 A0A1Q3FA94 A0A182FCZ4 K1QI70 A0A182PJ24 A0A182RQE1 Q17B59 A0A1S4G7L3 Q7QI48 A0A182UW62 A0A182WRZ8 A0A182ICX1 A0A1L8DTY0 A0A182KVW8 A0A182JW43 A0A182QRU2 A0A182MDL1 C3Z2Q1 A0A182NN59 A0A232FDX4 A0A293M3Q0 A0A1L8DTN8 K7IW26 A0A1Z5LGI1 A0A182HD29 A0A182SR45 A0A2R5L7E3 A0A182WK39 T1JIH3 Q4RXV6 H3DAT4 V9KHU4 H2TVC3 G3N6P5 A0A3B5KIU4 A0A3Q4MZ43 A0A3P9P9B6 A0A3P8USH6 A0A131XY44 B7PWV7 A0A3B5MQG3 A0A3B3QYH1 W5MZ09 A0A1B6E425 A0A1W5A8W1 A0A154PMJ9 A7S6W7 A0A3P8TFF6 A0A3B3QXW8 A0A3Q1HNM0 A0A3Q1CAZ9 A0A0L8HQK9 R7TQ36 A0A3Q0TBH3 A0A3Q3QXI8 A0A0L8HQR6 A0A0S7GTZ3 A0A3B3X2F1 A0A3B5QUV4 I3KS87 A0A3Q2CF56 A0A3B4F8U9 A0A3Q2WK80 A0A3P9BQM6
A0A195CE96 F4WYQ7 A0A151JZ65 A0A158N947 A0A026W788 A0A3L8DJ29 E2AK45 A0A151X4N5 A0A182ITC4 A0A336K518 Q17B57 A0A195BVM7 A0A182HD31 A0A084VXM2 A0A2A3EFL3 A0A088A4J1 B0W9N6 A0A210QI98 A0A0N0BFL6 A0A1Q3G4B2 Q17B58 E2C926 W5JFG0 A0A182TGD6 A0A1Q3FA94 A0A182FCZ4 K1QI70 A0A182PJ24 A0A182RQE1 Q17B59 A0A1S4G7L3 Q7QI48 A0A182UW62 A0A182WRZ8 A0A182ICX1 A0A1L8DTY0 A0A182KVW8 A0A182JW43 A0A182QRU2 A0A182MDL1 C3Z2Q1 A0A182NN59 A0A232FDX4 A0A293M3Q0 A0A1L8DTN8 K7IW26 A0A1Z5LGI1 A0A182HD29 A0A182SR45 A0A2R5L7E3 A0A182WK39 T1JIH3 Q4RXV6 H3DAT4 V9KHU4 H2TVC3 G3N6P5 A0A3B5KIU4 A0A3Q4MZ43 A0A3P9P9B6 A0A3P8USH6 A0A131XY44 B7PWV7 A0A3B5MQG3 A0A3B3QYH1 W5MZ09 A0A1B6E425 A0A1W5A8W1 A0A154PMJ9 A7S6W7 A0A3P8TFF6 A0A3B3QXW8 A0A3Q1HNM0 A0A3Q1CAZ9 A0A0L8HQK9 R7TQ36 A0A3Q0TBH3 A0A3Q3QXI8 A0A0L8HQR6 A0A0S7GTZ3 A0A3B3X2F1 A0A3B5QUV4 I3KS87 A0A3Q2CF56 A0A3B4F8U9 A0A3Q2WK80 A0A3P9BQM6
PDB
4CI3
E-value=6.68572e-33,
Score=352
Ontologies
GO
Topology
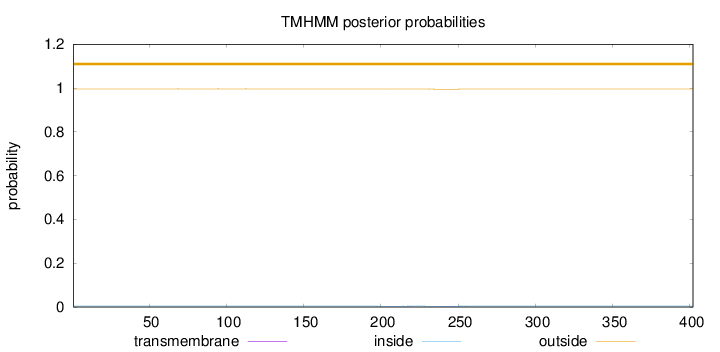
Length:
402
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04606
Exp number, first 60 AAs:
0.00022
Total prob of N-in:
0.00401
outside
1 - 402
Population Genetic Test Statistics
Pi
197.973886
Theta
173.86379
Tajima's D
-1.014713
CLR
1232.879397
CSRT
0.137443127843608
Interpretation
Uncertain
