Pre Gene Modal
BGIBMGA009383
Annotation
PREDICTED:_vacuolar_protein_sorting_26_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.124
Sequence
CDS
ATGTCTATAAATCTCTCTATATGCTTAAAACGTGCAAGTAAAATATATCACGAAGGGGAAATAATCGCTGGCGTGGTGGTGGTGGAGAGCAGTAGCGATGTGCGACATGAGGGCCTTTCACTCACTATGGAGGGATGTGTCAACTTGCAGCTGAGCACCAAGAATGTGGGCATATTTGAAGCTTTTTCAAATAGCATTAAGCCAATAAATTTAATAAATGTTACAGTTGAGCTAGCTCTCCCAGGTAAGATCCCTGTAGGCATCACAGAGATTCCATTTGAGATGCCACTGCGGGCCCGTCAGGCTGTATCTCCAGGATATCCTGGTCTACTGGAGACTTACCATGGAGTGTTTGTTAATATAATGTACACACTAAAGTGTAATATGAAGAGATCATTTCTCAATAAGCCACTGTTCACCACTTGCCAGTTCTTTGTACAGTATAGACATCAAGAACGCCCAGTACAGAAGGGTGTGCGGTGCGAGATGTCCGGTGCGTCGGTGCGCGCTGCGGGCACAGTGCCGCACTTCTCTGTGTTCGCAGATCTCACCTCCACCGTCTGCGCGTTGGATGCACCTGTCACTGGAAAGATCAGAGTTGACGAGTGCTCGGTACCCATCAAGTCCATTGAACTACAATTAGTGCGCGTTGAGACATGCGGCTGCGCTGACGGCTACTCTAGGGATGCAACCGAAATCCAGAATATCCAGATAGGGGAGGGAGACGTGGTGCGCGGCCGAGACATCCCGCTGTACATGGTGCTGCCGCGACTCTTCACTTGCCCCACCACCACCACGCTCAACTTTAAAATAGAGTTTGAACTGAACATCGCCGTGATATTTGAAGACGACTACTTGGTTACAGAAAATTTTCCGATATTACTTCTAAGGTGCAAGTAG
Protein
MSINLSICLKRASKIYHEGEIIAGVVVVESSSDVRHEGLSLTMEGCVNLQLSTKNVGIFEAFSNSIKPINLINVTVELALPGKIPVGITEIPFEMPLRARQAVSPGYPGLLETYHGVFVNIMYTLKCNMKRSFLNKPLFTTCQFFVQYRHQERPVQKGVRCEMSGASVRAAGTVPHFSVFADLTSTVCALDAPVTGKIRVDECSVPIKSIELQLVRVETCGCADGYSRDATEIQNIQIGEGDVVRGRDIPLYMVLPRLFTCPTTTTLNFKIEFELNIAVIFEDDYLVTENFPILLLRCK
Summary
Uniprot
Q2F5U5
H9JIN3
A0A2H1X1J7
A0A2A4K3N3
A0A2A4K2J9
S4PDS2
+ More
A0A194Q730 A0A212FFT3 A0A194RB22 A0A2A4K2X2 A0A0L7KU73 A0A2J7R457 K7ISC1 A0A232FDY8 A0A067QYX5 A0A1W5ACZ0 A0A158NSE5 A0A151HZ03 A0A0J7L5U6 W5MC60 E2AX49 R7U226 A0A1B6F1V8 A0A1B6J7C1 A0A195ES47 A0A151WMI1 Q6DHL2 E3TEZ4 E3TDC7 A0A2A3EKV9 M3ZL23 A0A151IE25 A0A3B4DPY4 H2MGT3 A0A3B3TYH4 H2SWQ0 A0A0L7RC82 A0A3P9JKC3 A0A087XL13 A0A3P9A141 A0A3S4QC84 A0A0S7H395 W5KNE3 A0A1S3I9N6 A0A0M8ZQ98 A0A210QUP8 A0A3Q2PZE5 A0A2I4CP14 A0A1Y1LVE8 A0A3Q3ER83 A0A3B3SWW7 A0A1X7UE65 G1NNY7 A0A1Y1LPK3 A0A3B4XXJ7 A0A3B4T8F1 Q4FZN8 E0VPR8 A0A1A8QYM4 A0A3Q2Y5I9 D7EJE8 A0A3Q2CG14 A0A1V4J3S6 A0A1A8ML13 A0A226PJH4 E1BSI1 B5X5W3 A0A402F8P1 A0A3Q3MPV3 H0Z3F4 A0A1A8GFN4 A0A3B5B818 A0A1L8HCE2 A0A3L8DIP8 U3IWV7 A0A096NJ37 A0A2K5IB84 F6QKV9 A0A0T6B3J5 A0A1A8IIL4 A0A1A8D989 A0A1A8B5T9 A0A2G8KWE7 A0A218US34 G1P4W8 A0A2K6AYM5 A0A2K6KVA1 A0A2K5ZGU9 A0A2K6QSB0 A0A2K5KZT4 G7P107 G7MMS9 A0A1U7SH90 W4XNG2 A0A1E1XH15 A0A0Q3PG51 A0A1U7R9T7 I0FND0 A0A151MEL2 G3SB23
A0A194Q730 A0A212FFT3 A0A194RB22 A0A2A4K2X2 A0A0L7KU73 A0A2J7R457 K7ISC1 A0A232FDY8 A0A067QYX5 A0A1W5ACZ0 A0A158NSE5 A0A151HZ03 A0A0J7L5U6 W5MC60 E2AX49 R7U226 A0A1B6F1V8 A0A1B6J7C1 A0A195ES47 A0A151WMI1 Q6DHL2 E3TEZ4 E3TDC7 A0A2A3EKV9 M3ZL23 A0A151IE25 A0A3B4DPY4 H2MGT3 A0A3B3TYH4 H2SWQ0 A0A0L7RC82 A0A3P9JKC3 A0A087XL13 A0A3P9A141 A0A3S4QC84 A0A0S7H395 W5KNE3 A0A1S3I9N6 A0A0M8ZQ98 A0A210QUP8 A0A3Q2PZE5 A0A2I4CP14 A0A1Y1LVE8 A0A3Q3ER83 A0A3B3SWW7 A0A1X7UE65 G1NNY7 A0A1Y1LPK3 A0A3B4XXJ7 A0A3B4T8F1 Q4FZN8 E0VPR8 A0A1A8QYM4 A0A3Q2Y5I9 D7EJE8 A0A3Q2CG14 A0A1V4J3S6 A0A1A8ML13 A0A226PJH4 E1BSI1 B5X5W3 A0A402F8P1 A0A3Q3MPV3 H0Z3F4 A0A1A8GFN4 A0A3B5B818 A0A1L8HCE2 A0A3L8DIP8 U3IWV7 A0A096NJ37 A0A2K5IB84 F6QKV9 A0A0T6B3J5 A0A1A8IIL4 A0A1A8D989 A0A1A8B5T9 A0A2G8KWE7 A0A218US34 G1P4W8 A0A2K6AYM5 A0A2K6KVA1 A0A2K5ZGU9 A0A2K6QSB0 A0A2K5KZT4 G7P107 G7MMS9 A0A1U7SH90 W4XNG2 A0A1E1XH15 A0A0Q3PG51 A0A1U7R9T7 I0FND0 A0A151MEL2 G3SB23
Pubmed
19121390
23622113
26354079
22118469
26227816
20075255
+ More
28648823 24845553 21347285 20798317 23254933 23594743 20634964 23542700 17554307 21551351 25069045 25329095 28812685 28004739 29240929 20838655 20566863 18362917 19820115 24621616 15592404 20433749 20360741 27762356 30249741 23749191 20431018 29023486 21993624 25362486 22002653 17431167 25319552 28503490 28071753 22293439 22398555
28648823 24845553 21347285 20798317 23254933 23594743 20634964 23542700 17554307 21551351 25069045 25329095 28812685 28004739 29240929 20838655 20566863 18362917 19820115 24621616 15592404 20433749 20360741 27762356 30249741 23749191 20431018 29023486 21993624 25362486 22002653 17431167 25319552 28503490 28071753 22293439 22398555
EMBL
DQ311328
ABD36272.1
BABH01027276
ODYU01012290
SOQ58524.1
NWSH01000219
+ More
PCG78280.1 PCG78279.1 GAIX01003536 JAA89024.1 KQ459337 KPJ01352.1 AGBW02008766 OWR52594.1 KQ460436 KPJ14694.1 PCG78278.1 JTDY01005622 KOB66818.1 NEVH01007815 PNF35610.1 NNAY01000349 OXU28996.1 KK852820 KDR15577.1 ADTU01024876 KQ976730 KYM76473.1 LBMM01000637 KMQ97928.1 AHAT01003118 AHAT01003119 AHAT01003120 GL443520 EFN61999.1 AMQN01010754 KB308480 ELT97716.1 GECZ01025559 JAS44210.1 GECU01012633 JAS95073.1 KQ981993 KYN31053.1 KQ982942 KYQ49074.1 CU633885 BC075958 AAH75958.1 GU588927 ADO28880.1 GU588357 ADO28313.1 KZ288226 PBC31892.1 KQ977894 KYM98963.1 KQ414616 KOC68438.1 AYCK01024797 NCKU01008995 RWS01546.1 GBYX01445785 GBYX01445784 JAO35654.1 KQ435911 KOX68905.1 NEDP02001809 OWF52426.1 GEZM01050490 JAV75576.1 GEZM01050492 JAV75573.1 BC099318 AAH99318.1 DS235371 EEB15374.1 HAEH01013714 HAEI01017106 SBR98114.1 DS497704 EFA12716.1 LSYS01009367 OPJ66407.1 HAEF01016278 HAEG01010240 SBR57437.1 AWGT02000067 OXB79517.1 AADN05000841 BT046432 ACI66233.1 BDOT01000143 GCF52809.1 ABQF01050728 ABQF01050729 HAEB01012057 HAEC01002503 SBQ70580.1 CM004468 OCT93774.1 QOIP01000007 RLU20325.1 ADON01026841 ADON01026842 AHZZ02021497 AHZZ02021498 AAMC01116142 AAMC01116143 LJIG01016069 KRT81707.1 HAED01010023 HAEE01016360 SBQ96235.1 HADZ01019054 HAEA01002360 SBQ30840.1 HADY01024387 HAEJ01000604 SBP62872.1 MRZV01000333 PIK52336.1 MUZQ01000153 OWK56559.1 AAPE02043825 AQIA01044460 AQIA01044461 CM001278 EHH51871.1 JSUE03027124 JU322030 JU332351 JU332352 JU473631 CM001255 AFE65786.1 AFH30435.1 EHH16976.1 AAGJ04141463 AAGJ04141464 GFAC01000822 JAT98366.1 LMAW01002568 KQK79723.1 JV045882 JV045883 JV045884 JV045885 JV045886 AFI35956.1 AKHW03006231 KYO22830.1 CABD030119306
PCG78280.1 PCG78279.1 GAIX01003536 JAA89024.1 KQ459337 KPJ01352.1 AGBW02008766 OWR52594.1 KQ460436 KPJ14694.1 PCG78278.1 JTDY01005622 KOB66818.1 NEVH01007815 PNF35610.1 NNAY01000349 OXU28996.1 KK852820 KDR15577.1 ADTU01024876 KQ976730 KYM76473.1 LBMM01000637 KMQ97928.1 AHAT01003118 AHAT01003119 AHAT01003120 GL443520 EFN61999.1 AMQN01010754 KB308480 ELT97716.1 GECZ01025559 JAS44210.1 GECU01012633 JAS95073.1 KQ981993 KYN31053.1 KQ982942 KYQ49074.1 CU633885 BC075958 AAH75958.1 GU588927 ADO28880.1 GU588357 ADO28313.1 KZ288226 PBC31892.1 KQ977894 KYM98963.1 KQ414616 KOC68438.1 AYCK01024797 NCKU01008995 RWS01546.1 GBYX01445785 GBYX01445784 JAO35654.1 KQ435911 KOX68905.1 NEDP02001809 OWF52426.1 GEZM01050490 JAV75576.1 GEZM01050492 JAV75573.1 BC099318 AAH99318.1 DS235371 EEB15374.1 HAEH01013714 HAEI01017106 SBR98114.1 DS497704 EFA12716.1 LSYS01009367 OPJ66407.1 HAEF01016278 HAEG01010240 SBR57437.1 AWGT02000067 OXB79517.1 AADN05000841 BT046432 ACI66233.1 BDOT01000143 GCF52809.1 ABQF01050728 ABQF01050729 HAEB01012057 HAEC01002503 SBQ70580.1 CM004468 OCT93774.1 QOIP01000007 RLU20325.1 ADON01026841 ADON01026842 AHZZ02021497 AHZZ02021498 AAMC01116142 AAMC01116143 LJIG01016069 KRT81707.1 HAED01010023 HAEE01016360 SBQ96235.1 HADZ01019054 HAEA01002360 SBQ30840.1 HADY01024387 HAEJ01000604 SBP62872.1 MRZV01000333 PIK52336.1 MUZQ01000153 OWK56559.1 AAPE02043825 AQIA01044460 AQIA01044461 CM001278 EHH51871.1 JSUE03027124 JU322030 JU332351 JU332352 JU473631 CM001255 AFE65786.1 AFH30435.1 EHH16976.1 AAGJ04141463 AAGJ04141464 GFAC01000822 JAT98366.1 LMAW01002568 KQK79723.1 JV045882 JV045883 JV045884 JV045885 JV045886 AFI35956.1 AKHW03006231 KYO22830.1 CABD030119306
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000037510
+ More
UP000235965 UP000002358 UP000215335 UP000027135 UP000192224 UP000005205 UP000078540 UP000036403 UP000018468 UP000000311 UP000014760 UP000078541 UP000075809 UP000000437 UP000221080 UP000242457 UP000002852 UP000078542 UP000261440 UP000001038 UP000261500 UP000005226 UP000053825 UP000265200 UP000028760 UP000265140 UP000285301 UP000018467 UP000085678 UP000053105 UP000242188 UP000265000 UP000192220 UP000261660 UP000261540 UP000007879 UP000001645 UP000261360 UP000261420 UP000009046 UP000264820 UP000007266 UP000265020 UP000190648 UP000198419 UP000000539 UP000087266 UP000288954 UP000261640 UP000007754 UP000261400 UP000186698 UP000279307 UP000016666 UP000028761 UP000233080 UP000008143 UP000230750 UP000197619 UP000001074 UP000233120 UP000233180 UP000233140 UP000233200 UP000233060 UP000009130 UP000233100 UP000006718 UP000189705 UP000007110 UP000051836 UP000189706 UP000050525 UP000001519
UP000235965 UP000002358 UP000215335 UP000027135 UP000192224 UP000005205 UP000078540 UP000036403 UP000018468 UP000000311 UP000014760 UP000078541 UP000075809 UP000000437 UP000221080 UP000242457 UP000002852 UP000078542 UP000261440 UP000001038 UP000261500 UP000005226 UP000053825 UP000265200 UP000028760 UP000265140 UP000285301 UP000018467 UP000085678 UP000053105 UP000242188 UP000265000 UP000192220 UP000261660 UP000261540 UP000007879 UP000001645 UP000261360 UP000261420 UP000009046 UP000264820 UP000007266 UP000265020 UP000190648 UP000198419 UP000000539 UP000087266 UP000288954 UP000261640 UP000007754 UP000261400 UP000186698 UP000279307 UP000016666 UP000028761 UP000233080 UP000008143 UP000230750 UP000197619 UP000001074 UP000233120 UP000233180 UP000233140 UP000233200 UP000233060 UP000009130 UP000233100 UP000006718 UP000189705 UP000007110 UP000051836 UP000189706 UP000050525 UP000001519
Pfam
PF03643 Vps26
SUPFAM
SSF81296
SSF81296
Gene 3D
ProteinModelPortal
Q2F5U5
H9JIN3
A0A2H1X1J7
A0A2A4K3N3
A0A2A4K2J9
S4PDS2
+ More
A0A194Q730 A0A212FFT3 A0A194RB22 A0A2A4K2X2 A0A0L7KU73 A0A2J7R457 K7ISC1 A0A232FDY8 A0A067QYX5 A0A1W5ACZ0 A0A158NSE5 A0A151HZ03 A0A0J7L5U6 W5MC60 E2AX49 R7U226 A0A1B6F1V8 A0A1B6J7C1 A0A195ES47 A0A151WMI1 Q6DHL2 E3TEZ4 E3TDC7 A0A2A3EKV9 M3ZL23 A0A151IE25 A0A3B4DPY4 H2MGT3 A0A3B3TYH4 H2SWQ0 A0A0L7RC82 A0A3P9JKC3 A0A087XL13 A0A3P9A141 A0A3S4QC84 A0A0S7H395 W5KNE3 A0A1S3I9N6 A0A0M8ZQ98 A0A210QUP8 A0A3Q2PZE5 A0A2I4CP14 A0A1Y1LVE8 A0A3Q3ER83 A0A3B3SWW7 A0A1X7UE65 G1NNY7 A0A1Y1LPK3 A0A3B4XXJ7 A0A3B4T8F1 Q4FZN8 E0VPR8 A0A1A8QYM4 A0A3Q2Y5I9 D7EJE8 A0A3Q2CG14 A0A1V4J3S6 A0A1A8ML13 A0A226PJH4 E1BSI1 B5X5W3 A0A402F8P1 A0A3Q3MPV3 H0Z3F4 A0A1A8GFN4 A0A3B5B818 A0A1L8HCE2 A0A3L8DIP8 U3IWV7 A0A096NJ37 A0A2K5IB84 F6QKV9 A0A0T6B3J5 A0A1A8IIL4 A0A1A8D989 A0A1A8B5T9 A0A2G8KWE7 A0A218US34 G1P4W8 A0A2K6AYM5 A0A2K6KVA1 A0A2K5ZGU9 A0A2K6QSB0 A0A2K5KZT4 G7P107 G7MMS9 A0A1U7SH90 W4XNG2 A0A1E1XH15 A0A0Q3PG51 A0A1U7R9T7 I0FND0 A0A151MEL2 G3SB23
A0A194Q730 A0A212FFT3 A0A194RB22 A0A2A4K2X2 A0A0L7KU73 A0A2J7R457 K7ISC1 A0A232FDY8 A0A067QYX5 A0A1W5ACZ0 A0A158NSE5 A0A151HZ03 A0A0J7L5U6 W5MC60 E2AX49 R7U226 A0A1B6F1V8 A0A1B6J7C1 A0A195ES47 A0A151WMI1 Q6DHL2 E3TEZ4 E3TDC7 A0A2A3EKV9 M3ZL23 A0A151IE25 A0A3B4DPY4 H2MGT3 A0A3B3TYH4 H2SWQ0 A0A0L7RC82 A0A3P9JKC3 A0A087XL13 A0A3P9A141 A0A3S4QC84 A0A0S7H395 W5KNE3 A0A1S3I9N6 A0A0M8ZQ98 A0A210QUP8 A0A3Q2PZE5 A0A2I4CP14 A0A1Y1LVE8 A0A3Q3ER83 A0A3B3SWW7 A0A1X7UE65 G1NNY7 A0A1Y1LPK3 A0A3B4XXJ7 A0A3B4T8F1 Q4FZN8 E0VPR8 A0A1A8QYM4 A0A3Q2Y5I9 D7EJE8 A0A3Q2CG14 A0A1V4J3S6 A0A1A8ML13 A0A226PJH4 E1BSI1 B5X5W3 A0A402F8P1 A0A3Q3MPV3 H0Z3F4 A0A1A8GFN4 A0A3B5B818 A0A1L8HCE2 A0A3L8DIP8 U3IWV7 A0A096NJ37 A0A2K5IB84 F6QKV9 A0A0T6B3J5 A0A1A8IIL4 A0A1A8D989 A0A1A8B5T9 A0A2G8KWE7 A0A218US34 G1P4W8 A0A2K6AYM5 A0A2K6KVA1 A0A2K5ZGU9 A0A2K6QSB0 A0A2K5KZT4 G7P107 G7MMS9 A0A1U7SH90 W4XNG2 A0A1E1XH15 A0A0Q3PG51 A0A1U7R9T7 I0FND0 A0A151MEL2 G3SB23
PDB
5F0J
E-value=4.9916e-07,
Score=127
Ontologies
PANTHER
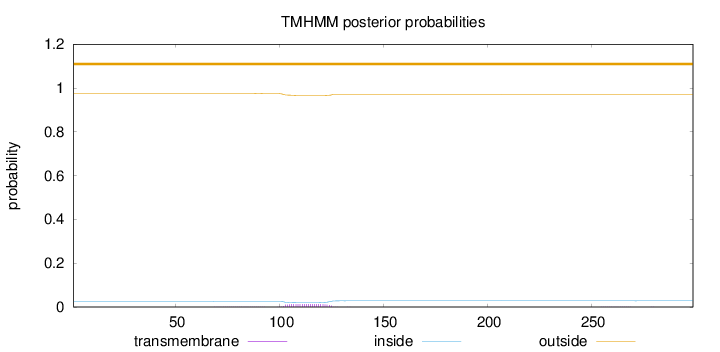
Topology
Length:
299
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.319969999999999
Exp number, first 60 AAs:
0.00076
Total prob of N-in:
0.02564
outside
1 - 299
Population Genetic Test Statistics
Pi
23.121239
Theta
37.217692
Tajima's D
-2.399441
CLR
0.659561
CSRT
0.00154992250387481
Interpretation
Uncertain
