Pre Gene Modal
BGIBMGA009382
Annotation
ligatin_[Danaus_plexippus]
Full name
Eukaryotic translation initiation factor 2D
Alternative Name
Ligatin
Location in the cell
Cytoplasmic Reliability : 1.566 Mitochondrial Reliability : 1.846
Sequence
CDS
ATGTTCGCAAAACCGTATAAGTTGAAATCCAATAACACGTTAAAGAATTCAGAAAAGAAACAGTTAGCGCAAAGGATCATTCATGAGTTTCCGACAATAACTGAAGAGAAAGTGAAGCAGCTGGTACCCGCCAAATCGATAGGCATTTGCATGAAATTGGTGTTGTCCTCTGGAGACATTGTAAATGTGTACGTGATAGACGGGGTGCCAATTATAATGGAAGTAGCAGAAGGGTTGGTCCCGACGGTGTGCGCGCTCTGGCAAGCTCCTGAAATGGTCCCACATATCATTATACATACACCGGTCTTTCCAAAGGTCCAAGGTGGAGCTCCTCTATACCTCCCAGGAGTAGAACTACCAGCAGGAGGTACAGGTTTCCCACAGTTTTGTAAAGGTGAATTAATTGCAGCACATACTAGTAATAATTCTGCCACAGTTATTGTTGGTCGAGCTATTCTTTCCAGCGGAGACATGCTACTGAGAACTGCTGGAGTATGCATGGAGACACTTCATGTATTTGGGGATCATTTAAGCAAAGATATAAAATTTGGTAAACTTGAACGTCCGAAGCTTGGACCTCCTGCATATGGTGGTGGCAATGCAGCTCAAAACTTGGCCAACAATATACAGAAGTTAAGCATACAACCAGCTGTAAAGGAGGAATGGCCTGCACTCGGCCAGCCGCCTGCTCCTGTACCAAACGAACCACAACTGATACCTGAACCACCGAACCCAGTAGTTGATACACCTACCCAATTAGAGGATGGAAACTGTTTAGCCATAGATTCCGATGAGGAAGATAATGTGCCTACCGACATGGACGGGTTGCTGAGGTGGTGCATGCTGTCCTTCCTCAAGCTGGAAGGGAACAAGGTCGAACTGCCGCTCAAAACCAACCTGCTCTACAGAAATCATCTGATGGCTCTGTGCCCGGCGGAGCGGACGCTGGACGTCAAAAAGTCATCCTTTAAGAAAATGGGGAAGTTTTTAGAGGCCATGGAAAAGGAAGGCCTGGTCCGAGTCTGCGAGCTGGAGAAGGGCGTGGACGCCGTCGTGCAGCTGTCCCCCTCCCACCCCGCCGTGCGCGCGCACCCGCCCCCCCGCGCGCCCGCCCGCCCCGCCCCCCGCGCCGCGCCCCCCGCCCCCCCGCGGCTCCGCGAGGTGCAGTGCGTGACGGCGGCCGTGGCGCCGCTGTTCGCCCCCCTCAAGAAGGGCACCGCGCTGACACCCGCGGACGTCCGCGCGACCCTGACGGGGCACGTGAGGTCCGCGGGGCTCGCTGTCCCGGGGCAGAGCGCCGTCCGCCTCGACCTCACGCTGGCCACAGTGTTGGGTCGGCAGCCCGGGGAACACCTAACGTGGGAGGAGCTGATGTCGGGTGTTCTGAAGAAAATGACTCCGAGTACTGAGATACAGTTCGCCGACGGAGAGCTGAGGCTGTCCAGGAGCCGCCTCGAGCCCGTCACCATGCAGGTCGCCAGCCGCAGCGGAAACAAAAAGGTGACTCTAGTGTCTAACCTGGAAGCATATGGATTCAACCTGTTGGAGCTGAGCGCAGTTTGTCAGCGAGGAGTGGCAGCCTCCTGCGGCGTCACGCGCTCTCCTGGCGCTAAACACGACCAGCTCATGCTGCAAGGAGACCAGACACATTTTGTTGCTAAACTACTGATCGAAAAATATGGACTGCCAAAAAAATTTGTAGAAGGTGCTGACAAGGCATTAAACAGGAAGAAGTAG
Protein
MFAKPYKLKSNNTLKNSEKKQLAQRIIHEFPTITEEKVKQLVPAKSIGICMKLVLSSGDIVNVYVIDGVPIIMEVAEGLVPTVCALWQAPEMVPHIIIHTPVFPKVQGGAPLYLPGVELPAGGTGFPQFCKGELIAAHTSNNSATVIVGRAILSSGDMLLRTAGVCMETLHVFGDHLSKDIKFGKLERPKLGPPAYGGGNAAQNLANNIQKLSIQPAVKEEWPALGQPPAPVPNEPQLIPEPPNPVVDTPTQLEDGNCLAIDSDEEDNVPTDMDGLLRWCMLSFLKLEGNKVELPLKTNLLYRNHLMALCPAERTLDVKKSSFKKMGKFLEAMEKEGLVRVCELEKGVDAVVQLSPSHPAVRAHPPPRAPARPAPRAAPPAPPRLREVQCVTAAVAPLFAPLKKGTALTPADVRATLTGHVRSAGLAVPGQSAVRLDLTLATVLGRQPGEHLTWEELMSGVLKKMTPSTEIQFADGELRLSRSRLEPVTMQVASRSGNKKVTLVSNLEAYGFNLLELSAVCQRGVAASCGVTRSPGAKHDQLMLQGDQTHFVAKLLIEKYGLPKKFVEGADKALNRKK
Summary
Description
Translation initiation factor that is able to deliver tRNA to the P-site of the eukaryotic ribosome in a GTP-independent manner. The binding of Met-tRNA(I) occurs after the AUG codon finds its position in the P-site of 40S ribosomes, the situation that takes place during initiation complex formation on some specific RNAs. Its activity in tRNA binding with 40S subunits does not require the presence of the aminoacyl moiety. Possesses the unique ability to deliver non-Met (elongator) tRNAs into the P-site of the 40S subunit. In addition to its role in initiation, can promote release of deacylated tRNA and mRNA from recycled 40S subunits following ABCE1-mediated dissociation of post-termination ribosomal complexes into subunits.
Translation initiation factor that is able to deliver tRNA to the P-site of the eukaryotic ribosome in a GTP-independent manner. The binding of Met-tRNA(I) occurs after the AUG codon finds its position in the P-site of 40S ribosomes, the situation that takes place during initiation complex formation on some specific RNAs. Its activity in tRNA binding with 40S subunits does not require the presence of the aminoacyl moiety. Possesses the unique ability to deliver non-Met (elongator) tRNAs into the P-site of the 40S subunit. In addition to its role in initiation, can promote release of deacylated tRNA and mRNA from recycled 40S subunits following ABCE1-mediated dissociation of post-termination ribosomal complexes into subunits (By similarity).
Translation initiation factor that is able to deliver tRNA to the P-site of the eukaryotic ribosome in a GTP-independent manner. The binding of Met-tRNA(I) occurs after the AUG codon finds its position in the P-site of 40S ribosomes, the situation that takes place during initiation complex formation on some specific RNAs. Its activity in tRNA binding with 40S subunits does not require the presence of the aminoacyl moiety. Possesses the unique ability to deliver non-Met (elongator) tRNAs into the P-site of the 40S subunit. In addition to its role in initiation, can promote release of deacylated tRNA and mRNA from recycled 40S subunits following ABCE1-mediated dissociation of post-termination ribosomal complexes into subunits (By similarity).
Similarity
Belongs to the eIF2D family.
Keywords
Acetylation
Complete proteome
Cytoplasm
Initiation factor
Phosphoprotein
Protein biosynthesis
Reference proteome
Feature
chain Eukaryotic translation initiation factor 2D
Uniprot
H9JIN2
A0A2H1VTH1
A0A2A4K3D8
S4PFS5
A0A212FFS6
A0A194Q7J5
+ More
A0A437B689 A0A194RB28 A0A067QYP9 A0A2J7PXW8 A0A2P8XTV6 E9GLT4 N6U6J2 U4TZQ4 A0A0P6DA09 A0A164XR31 A0A139WNU2 A0A0P6C1P9 A0A0P6JJC7 A0A232FC55 D7EIK7 A0A0P6AL47 K7IPW9 A0A158NDW2 A0A1W4WQ12 A0A1W4X109 A0A452SB30 A0A195BR84 A0A151X4C3 A0A3Q7UJD6 A0A195ESA2 A0A087ZTQ3 A0A0L7RBI1 A0A0T6AU92 A0A0J7KFV6 F4W7V5 C3ZJ25 A0A195C4Z6 W5N343 A0A1S3FAH5 K9IMH5 A0A3Q7RYA2 K9ILP3 A0A195D7W7 A0A452SB07 E2AXJ4 A0A3Q7V015 P0CL18 A0A452E064 H0VDH5 Q5PPG7 A0A452E018 A0A2Y9K462 A0A154PLU8 B4NBG8 A0A1B6CUD5 A0A1A8F650 M3YKZ0 A0A2A3EBV3 G5BP24 E9IGR3 E2C5I3 A0A452DZP7 A0A1W4U5D6 A0A250Y350 A0A2Y9DIX6 A7RTR9 A0A3L8DGZ3 A0A3B3SN44 A0A0M3QXT1 A0A3R7QX09 A0A2R9AWK3 K7C945 Q58CR3 G7MER3 A0A2Y9JU02 G3RGZ7 L8IVZ3 H2N3X8 B4KAZ0 A0A0M8ZVG6 A0A3M6TFL1 A0A1A8V8U8 G3SLV4 L5JXR6 K7F6S9 Q9NG60 W5P7E3 G8JKW9 A0A1A8EIN3 A0A034VV88 G1QIW0 A0A437D6E0 A0A3B3BVD3 Q9GU72 B4PQ62 A0A1A8JS25 A0A0L8H398 Q29AB1
A0A437B689 A0A194RB28 A0A067QYP9 A0A2J7PXW8 A0A2P8XTV6 E9GLT4 N6U6J2 U4TZQ4 A0A0P6DA09 A0A164XR31 A0A139WNU2 A0A0P6C1P9 A0A0P6JJC7 A0A232FC55 D7EIK7 A0A0P6AL47 K7IPW9 A0A158NDW2 A0A1W4WQ12 A0A1W4X109 A0A452SB30 A0A195BR84 A0A151X4C3 A0A3Q7UJD6 A0A195ESA2 A0A087ZTQ3 A0A0L7RBI1 A0A0T6AU92 A0A0J7KFV6 F4W7V5 C3ZJ25 A0A195C4Z6 W5N343 A0A1S3FAH5 K9IMH5 A0A3Q7RYA2 K9ILP3 A0A195D7W7 A0A452SB07 E2AXJ4 A0A3Q7V015 P0CL18 A0A452E064 H0VDH5 Q5PPG7 A0A452E018 A0A2Y9K462 A0A154PLU8 B4NBG8 A0A1B6CUD5 A0A1A8F650 M3YKZ0 A0A2A3EBV3 G5BP24 E9IGR3 E2C5I3 A0A452DZP7 A0A1W4U5D6 A0A250Y350 A0A2Y9DIX6 A7RTR9 A0A3L8DGZ3 A0A3B3SN44 A0A0M3QXT1 A0A3R7QX09 A0A2R9AWK3 K7C945 Q58CR3 G7MER3 A0A2Y9JU02 G3RGZ7 L8IVZ3 H2N3X8 B4KAZ0 A0A0M8ZVG6 A0A3M6TFL1 A0A1A8V8U8 G3SLV4 L5JXR6 K7F6S9 Q9NG60 W5P7E3 G8JKW9 A0A1A8EIN3 A0A034VV88 G1QIW0 A0A437D6E0 A0A3B3BVD3 Q9GU72 B4PQ62 A0A1A8JS25 A0A0L8H398 Q29AB1
Pubmed
19121390
23622113
22118469
26354079
24845553
29403074
+ More
21292972 23537049 18362917 19820115 28648823 20075255 21347285 21719571 18563158 20798317 20713520 21993624 15489334 22673903 17994087 21993625 21282665 28087693 17615350 30249741 29240929 22722832 16305752 22002653 22398555 22751099 30382153 23258410 17381049 10731132 11157774 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20809919 19393038 25348373 29451363 17550304 15632085
21292972 23537049 18362917 19820115 28648823 20075255 21347285 21719571 18563158 20798317 20713520 21993624 15489334 22673903 17994087 21993625 21282665 28087693 17615350 30249741 29240929 22722832 16305752 22002653 22398555 22751099 30382153 23258410 17381049 10731132 11157774 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20809919 19393038 25348373 29451363 17550304 15632085
EMBL
BABH01027274
ODYU01004328
SOQ44078.1
NWSH01000219
PCG78283.1
GAIX01003897
+ More
JAA88663.1 AGBW02008766 OWR52591.1 KQ459337 KPJ01359.1 RSAL01000154 RVE45673.1 KQ460436 KPJ14699.1 KK852819 KDR15625.1 NEVH01020852 PNF21188.1 PYGN01001358 PSN35430.1 GL732551 EFX79561.1 APGK01040754 KB740984 ENN76271.1 KB631904 ERL87099.1 GDIQ01081144 JAN13593.1 LRGB01000944 KZS14484.1 KQ971307 KYB29678.1 GDIP01008131 JAM95584.1 GDIQ01005823 JAN88914.1 NNAY01000436 OXU28401.1 EFA12044.2 GDIP01041307 JAM62408.1 ADTU01013048 ADTU01013049 ADTU01013050 KQ976424 KYM88471.1 KQ982548 KYQ55272.1 KQ981993 KYN30782.1 KQ414617 KOC68198.1 LJIG01022783 KRT78739.1 LBMM01008077 KMQ89162.1 GL887888 EGI69625.1 GG666631 EEN47454.1 KQ978251 KYM95944.1 AHAT01016655 AHAT01016656 GABZ01005188 JAA48337.1 GABZ01005187 JAA48338.1 KQ981153 KYN08966.1 GL443590 EFN61832.1 LWLT01000016 AAKN02017887 BC087701 KQ434977 KZC12822.1 CH964232 EDW81132.1 GEDC01020264 JAS17034.1 HAEB01008060 HAEC01015932 SBQ54587.1 AEYP01041303 AEYP01041304 KZ288291 PBC29215.1 JH171215 EHB11035.1 GL763054 EFZ20228.1 GL452770 EFN76796.1 GFFV01001922 JAV38023.1 DS469538 EDO45078.1 QOIP01000008 RLU19591.1 CP012526 ALC46409.1 QCYY01000993 ROT81300.1 AJFE02080111 GABF01006718 JAA15427.1 BT021884 BC114158 CM001253 EHH15610.1 CABD030008537 CABD030008538 JH880730 ELR59317.1 ABGA01259996 ABGA01259997 ABGA01259998 NDHI03003431 PNJ53237.1 CH933806 EDW14668.1 KQ435851 KOX70836.1 RCHS01003687 RMX40099.1 HADY01003631 HAEJ01016323 SBS56780.1 KB031109 ELK03296.1 AGCU01190819 AGCU01190820 AGCU01190821 AF260235 AY051703 AE014297 AAF67203.1 AAK93127.1 AAN14162.1 AMGL01020019 AMGL01020020 HAEA01016464 SBQ44945.1 GAKP01013489 JAC45463.1 ADFV01074387 CM012443 RVE70725.1 AF186594 AAG16724.1 CM000160 EDW98331.1 HAED01017231 HAEE01002174 SBR22194.1 KQ419539 KOF83240.1 CM000070 EAL27439.1
JAA88663.1 AGBW02008766 OWR52591.1 KQ459337 KPJ01359.1 RSAL01000154 RVE45673.1 KQ460436 KPJ14699.1 KK852819 KDR15625.1 NEVH01020852 PNF21188.1 PYGN01001358 PSN35430.1 GL732551 EFX79561.1 APGK01040754 KB740984 ENN76271.1 KB631904 ERL87099.1 GDIQ01081144 JAN13593.1 LRGB01000944 KZS14484.1 KQ971307 KYB29678.1 GDIP01008131 JAM95584.1 GDIQ01005823 JAN88914.1 NNAY01000436 OXU28401.1 EFA12044.2 GDIP01041307 JAM62408.1 ADTU01013048 ADTU01013049 ADTU01013050 KQ976424 KYM88471.1 KQ982548 KYQ55272.1 KQ981993 KYN30782.1 KQ414617 KOC68198.1 LJIG01022783 KRT78739.1 LBMM01008077 KMQ89162.1 GL887888 EGI69625.1 GG666631 EEN47454.1 KQ978251 KYM95944.1 AHAT01016655 AHAT01016656 GABZ01005188 JAA48337.1 GABZ01005187 JAA48338.1 KQ981153 KYN08966.1 GL443590 EFN61832.1 LWLT01000016 AAKN02017887 BC087701 KQ434977 KZC12822.1 CH964232 EDW81132.1 GEDC01020264 JAS17034.1 HAEB01008060 HAEC01015932 SBQ54587.1 AEYP01041303 AEYP01041304 KZ288291 PBC29215.1 JH171215 EHB11035.1 GL763054 EFZ20228.1 GL452770 EFN76796.1 GFFV01001922 JAV38023.1 DS469538 EDO45078.1 QOIP01000008 RLU19591.1 CP012526 ALC46409.1 QCYY01000993 ROT81300.1 AJFE02080111 GABF01006718 JAA15427.1 BT021884 BC114158 CM001253 EHH15610.1 CABD030008537 CABD030008538 JH880730 ELR59317.1 ABGA01259996 ABGA01259997 ABGA01259998 NDHI03003431 PNJ53237.1 CH933806 EDW14668.1 KQ435851 KOX70836.1 RCHS01003687 RMX40099.1 HADY01003631 HAEJ01016323 SBS56780.1 KB031109 ELK03296.1 AGCU01190819 AGCU01190820 AGCU01190821 AF260235 AY051703 AE014297 AAF67203.1 AAK93127.1 AAN14162.1 AMGL01020019 AMGL01020020 HAEA01016464 SBQ44945.1 GAKP01013489 JAC45463.1 ADFV01074387 CM012443 RVE70725.1 AF186594 AAG16724.1 CM000160 EDW98331.1 HAED01017231 HAEE01002174 SBR22194.1 KQ419539 KOF83240.1 CM000070 EAL27439.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000283053
UP000053240
+ More
UP000027135 UP000235965 UP000245037 UP000000305 UP000019118 UP000030742 UP000076858 UP000007266 UP000215335 UP000002358 UP000005205 UP000192223 UP000291022 UP000078540 UP000075809 UP000286642 UP000078541 UP000005203 UP000053825 UP000036403 UP000007755 UP000001554 UP000078542 UP000018468 UP000081671 UP000286640 UP000078492 UP000000311 UP000001811 UP000291000 UP000005447 UP000002494 UP000248482 UP000076502 UP000007798 UP000000715 UP000242457 UP000006813 UP000008237 UP000192221 UP000248480 UP000001593 UP000279307 UP000261540 UP000092553 UP000283509 UP000240080 UP000009136 UP000001519 UP000001595 UP000009192 UP000053105 UP000275408 UP000007646 UP000010552 UP000007267 UP000000803 UP000002356 UP000001073 UP000261560 UP000002282 UP000053454 UP000001819
UP000027135 UP000235965 UP000245037 UP000000305 UP000019118 UP000030742 UP000076858 UP000007266 UP000215335 UP000002358 UP000005205 UP000192223 UP000291022 UP000078540 UP000075809 UP000286642 UP000078541 UP000005203 UP000053825 UP000036403 UP000007755 UP000001554 UP000078542 UP000018468 UP000081671 UP000286640 UP000078492 UP000000311 UP000001811 UP000291000 UP000005447 UP000002494 UP000248482 UP000076502 UP000007798 UP000000715 UP000242457 UP000006813 UP000008237 UP000192221 UP000248480 UP000001593 UP000279307 UP000261540 UP000092553 UP000283509 UP000240080 UP000009136 UP000001519 UP000001595 UP000009192 UP000053105 UP000275408 UP000007646 UP000010552 UP000007267 UP000000803 UP000002356 UP000001073 UP000261560 UP000002282 UP000053454 UP000001819
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JIN2
A0A2H1VTH1
A0A2A4K3D8
S4PFS5
A0A212FFS6
A0A194Q7J5
+ More
A0A437B689 A0A194RB28 A0A067QYP9 A0A2J7PXW8 A0A2P8XTV6 E9GLT4 N6U6J2 U4TZQ4 A0A0P6DA09 A0A164XR31 A0A139WNU2 A0A0P6C1P9 A0A0P6JJC7 A0A232FC55 D7EIK7 A0A0P6AL47 K7IPW9 A0A158NDW2 A0A1W4WQ12 A0A1W4X109 A0A452SB30 A0A195BR84 A0A151X4C3 A0A3Q7UJD6 A0A195ESA2 A0A087ZTQ3 A0A0L7RBI1 A0A0T6AU92 A0A0J7KFV6 F4W7V5 C3ZJ25 A0A195C4Z6 W5N343 A0A1S3FAH5 K9IMH5 A0A3Q7RYA2 K9ILP3 A0A195D7W7 A0A452SB07 E2AXJ4 A0A3Q7V015 P0CL18 A0A452E064 H0VDH5 Q5PPG7 A0A452E018 A0A2Y9K462 A0A154PLU8 B4NBG8 A0A1B6CUD5 A0A1A8F650 M3YKZ0 A0A2A3EBV3 G5BP24 E9IGR3 E2C5I3 A0A452DZP7 A0A1W4U5D6 A0A250Y350 A0A2Y9DIX6 A7RTR9 A0A3L8DGZ3 A0A3B3SN44 A0A0M3QXT1 A0A3R7QX09 A0A2R9AWK3 K7C945 Q58CR3 G7MER3 A0A2Y9JU02 G3RGZ7 L8IVZ3 H2N3X8 B4KAZ0 A0A0M8ZVG6 A0A3M6TFL1 A0A1A8V8U8 G3SLV4 L5JXR6 K7F6S9 Q9NG60 W5P7E3 G8JKW9 A0A1A8EIN3 A0A034VV88 G1QIW0 A0A437D6E0 A0A3B3BVD3 Q9GU72 B4PQ62 A0A1A8JS25 A0A0L8H398 Q29AB1
A0A437B689 A0A194RB28 A0A067QYP9 A0A2J7PXW8 A0A2P8XTV6 E9GLT4 N6U6J2 U4TZQ4 A0A0P6DA09 A0A164XR31 A0A139WNU2 A0A0P6C1P9 A0A0P6JJC7 A0A232FC55 D7EIK7 A0A0P6AL47 K7IPW9 A0A158NDW2 A0A1W4WQ12 A0A1W4X109 A0A452SB30 A0A195BR84 A0A151X4C3 A0A3Q7UJD6 A0A195ESA2 A0A087ZTQ3 A0A0L7RBI1 A0A0T6AU92 A0A0J7KFV6 F4W7V5 C3ZJ25 A0A195C4Z6 W5N343 A0A1S3FAH5 K9IMH5 A0A3Q7RYA2 K9ILP3 A0A195D7W7 A0A452SB07 E2AXJ4 A0A3Q7V015 P0CL18 A0A452E064 H0VDH5 Q5PPG7 A0A452E018 A0A2Y9K462 A0A154PLU8 B4NBG8 A0A1B6CUD5 A0A1A8F650 M3YKZ0 A0A2A3EBV3 G5BP24 E9IGR3 E2C5I3 A0A452DZP7 A0A1W4U5D6 A0A250Y350 A0A2Y9DIX6 A7RTR9 A0A3L8DGZ3 A0A3B3SN44 A0A0M3QXT1 A0A3R7QX09 A0A2R9AWK3 K7C945 Q58CR3 G7MER3 A0A2Y9JU02 G3RGZ7 L8IVZ3 H2N3X8 B4KAZ0 A0A0M8ZVG6 A0A3M6TFL1 A0A1A8V8U8 G3SLV4 L5JXR6 K7F6S9 Q9NG60 W5P7E3 G8JKW9 A0A1A8EIN3 A0A034VV88 G1QIW0 A0A437D6E0 A0A3B3BVD3 Q9GU72 B4PQ62 A0A1A8JS25 A0A0L8H398 Q29AB1
PDB
5OA3
E-value=1.62285e-62,
Score=609
Ontologies
GO
PANTHER
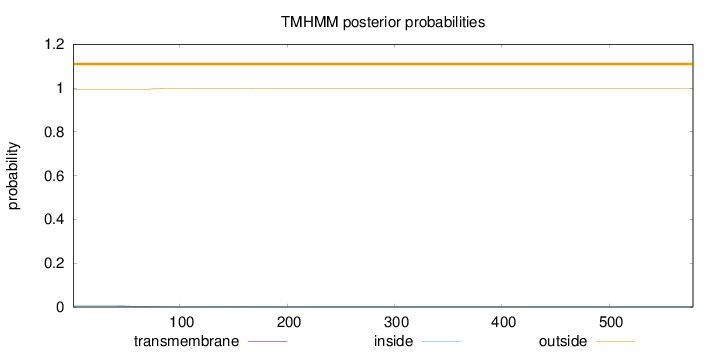
Topology
Subcellular location
Cytoplasm
Length:
578
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.11175
Exp number, first 60 AAs:
0.0405
Total prob of N-in:
0.00678
outside
1 - 578
Population Genetic Test Statistics
Pi
264.834042
Theta
213.605286
Tajima's D
0.423414
CLR
103.91952
CSRT
0.490425478726064
Interpretation
Uncertain
