Pre Gene Modal
BGIBMGA009312
Annotation
growth_arrest-specific_protein_2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.338
Sequence
CDS
ATGGCGCTGTATCGGTGCGGGCCGACTGCTCTGGGCAGCCGCACCTCCTGGGGCCCGGTGTCGAGTCCTGAACACAAACCCTTCCCGCGCCCTCTCAGTGGGAACTTTGACAGAATATTCAACGCTTTCGAAAGGACCAACGCCTTCGAGTACGTCCGAAGCAACTTGGACGGCAAGGAGGAGGCTCGCCATGTAGGTCATGCGGATGTAGTGGTGAACAGGAAGATGAGCAGCAGCTACGAAAGAGCCGACTGCGCGCTGGCTGCCCAGGCTCCTTGTGATGACGAAGACTTCTACAGAGAGAAGATCCTGTACTCGCAGGCGAGGCAGCTGTTTCCGCTGCAAGAGGACCTGGCTGACTGGATTAATAAGACTATTGGCATAACATACCTGACCGGAGAGAATTTTCTGGACGTGTTGGACAACGGCGTGGAGCTGTGTCAGCTGGCGAGCGTCATACATGAGCGAGCTCGGGGCGCGCTCGACCAGGGACTCGTTGTGGGGGTGGTGCCACAGATCCGCGGCCGCTGCTGGCAGCGCGCGGCCCGTCGCAGCTTCTTCTCCAGGGACAACACGGAGAACTTCATCACGTTCTGCCGCGAGCTTGGAGTCCACGAGAACCTGCTGTTCGAGAGCGATGACCTCGTTCTCCACAACCAACCCCGCCAAGTGATCCTCTGCCTGTTGGAAGTGGCTAGACTGGCCACCAAGTTCAACATAGAGCCGCCGGGGCTGGTACAGCTAGAGAAGGAGATCGCGATGGAAGAGCGCGACTCCGGCCTCGACTCGGCCATGTCTAGCGCTGCCTGGCAGTTCAGAGACAGCTCGCCCGAACCCGACAAAACTCTGGTTGAAAATTCGGCCCCAACCCCAGATCCAAATGAAAGCCAAGAGTCAGCGGTGGACAACGCCGACGCCGAGAGCACCAAGTCTGAGGGGACGGTGGGCAGCACTGACTCCGATGTGCCGCTCAAACCCACCAATGAACTCGATAAGAGAGTGCAACTCGTGACGCGGCTGATGGAACGCGGCTGCAGCTGTAACTCCGGGAAGTGTTCCAAGCTGCTCAAAGTGAAGAAGGTCGGAGAAGGACGCTACAACATCGCAGGAAGGAATGTCTTCATTCGGTTATTGAAGGGTCGCCACATGATGGTGCGTGTCGGCGGCGGCTGGGACACGCTGGAGCACTTCCTGTCGCGGCATGAACCGTGCCAGGTGCGCCTCGTCACGCAAGGACGCCGCGCATCAGTGCTGCCTGTACCGACCACACCAGCCTCGCCTGTCACACCCGTCACGCCCGGCACGCCCCCCTCACCAGTCACCCCCGCCTCACCCGCCGCGTCTAGCAAGCACAGCAGTGGACGTAGCTCCCCTCTTCCGCCTCCACCGCGGACGTCCACTCCACGACGTCCCGCACTGTCCCTGCCACTACGCTCCACCGCCCCGTCACCCGCCCCCTCGTTATCCCACAGCACGCCCTCCACGCCAATCTCGCGCTCGGTCCGTACTCCACCCGCAGAGTTCCGCAAGTCGTTGACCTCCGCAAACCTGCTCGGAGCGCCCAAGAAGTCCCGCAGCGCCAGCCTCGCGAGTGAAGGCGGTGGTGAGCGGTCCATCCGCAATAAGAAGAAGTCCGTGAACACCGTCGCTGCCTCGCCGGCGCCGCGCAAACTCCGCAGCCAGAGCTTGGTGCCGGCGGCTAGCGGAGCACCGGGCGCGGCGGGGGGTGCGGCGGGGAGCGCGGTGGCGGGCGCGGCGCGGAAGGCTCGCAGTCTCAGCGCGGCGAGCGGCGGCGCCCTCACGCAGGCGGCTATCGAGGAGAGCATCAGACTCTCCCTGGCGGCGAGCATGGTGGACGCAGAGTGCGCCAATAAGCCATTCCTCCACATCAAAGCGAAGTACCGCAGTCCACCGCCACGGGAGGTGCCTCCCAGATAA
Protein
MALYRCGPTALGSRTSWGPVSSPEHKPFPRPLSGNFDRIFNAFERTNAFEYVRSNLDGKEEARHVGHADVVVNRKMSSSYERADCALAAQAPCDDEDFYREKILYSQARQLFPLQEDLADWINKTIGITYLTGENFLDVLDNGVELCQLASVIHERARGALDQGLVVGVVPQIRGRCWQRAARRSFFSRDNTENFITFCRELGVHENLLFESDDLVLHNQPRQVILCLLEVARLATKFNIEPPGLVQLEKEIAMEERDSGLDSAMSSAAWQFRDSSPEPDKTLVENSAPTPDPNESQESAVDNADAESTKSEGTVGSTDSDVPLKPTNELDKRVQLVTRLMERGCSCNSGKCSKLLKVKKVGEGRYNIAGRNVFIRLLKGRHMMVRVGGGWDTLEHFLSRHEPCQVRLVTQGRRASVLPVPTTPASPVTPVTPGTPPSPVTPASPAASSKHSSGRSSPLPPPPRTSTPRRPALSLPLRSTAPSPAPSLSHSTPSTPISRSVRTPPAEFRKSLTSANLLGAPKKSRSASLASEGGGERSIRNKKKSVNTVAASPAPRKLRSQSLVPAASGAPGAAGGAAGSAVAGAARKARSLSAASGGALTQAAIEESIRLSLAASMVDAECANKPFLHIKAKYRSPPPREVPPR
Summary
Uniprot
Proteomes
PRIDE
Gene 3D
CDD
ProteinModelPortal
PDB
1V5R
E-value=1.63514e-09,
Score=152
Ontologies
GO
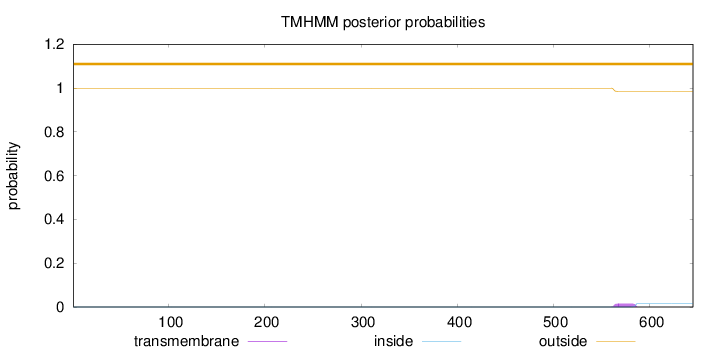
Topology
Length:
645
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.34803
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00043
outside
1 - 645
Population Genetic Test Statistics
Pi
209.849535
Theta
170.241937
Tajima's D
0.693159
CLR
0.625856
CSRT
0.568921553922304
Interpretation
Uncertain
