Pre Gene Modal
BGIBMGA009380
Annotation
PREDICTED:_septin-2-like_isoform_X1_[Amyelois_transitella]
Full name
Septin-2
Location in the cell
Nuclear Reliability : 4.12
Sequence
CDS
ATGGCGGCGATCGATGTTGAGCCAACTAAAGCGGAACCTACCCTACGCACCTTGAAACTTTCGGGACATGTTGGCTTCGACAGTCTACCTGACCAGCTGGTCAGTAAAAGCATACAGAATGGTTTCGTCTTCAATATCCTGTGTATAGGTGAAACTGGCCTTGGTAAATCAACTTTAATGGATTCTCTGTTCAATACCAACTTTGAATCATCTCCGAGTCCTCATAACTTACCTACTGTTAAGTTGAAGGCTCACACTTATGAGCTGGAGGAGAGCAATGTCAGGCTGAAGTTAACAATCTGTGACACTGTCGGTTACGGTGACCAAGTGAATAAAGAAGACAGTTTCAAAGCGGTGGTGGATTACATTGATGCACAGTTCGAGGCCTATCTCCAGGAGGAGCTCAAGATCAAGCGTTCACTGTCCAGTTATCATGACAGCAGACTGCATGTCTGTCTGTACTTCATCTGTCCAACAGGTCATGGTTTGAAGTCCATTGATTTGGTGTGTATGAAGAAACTCGATACCAAAGTGAACATCATACCGATCATCGCTAAGGCTGACACCATCTCCAAGACTGAGCTGCAGAGGTTCAAGTCCAAAATAATGCAAGAGCTGCAGTCGAACGGCGTGGAGATCTACCAGTTCCCGGTGGACGACGAGTCCGTGTCCGAGGTCAACGGCTCCATGAACTCGCACCTGCCCTTCGCCGTCGTCGGCAGCACCGACTTCGTGAAGATCGGCAACAAGACCGTGCGCGCGCGCCAGTACCCCTGGGGCACCGTGCAAGTGGAGAGCGAGTCCCACTGCGACTTCGTGAAGCTGCGCGAGATGCTGATCCGCACCAACATGGAGGACATGCGCGACAAGACGCACGCGCGCCACTACGAGCTGTACCGCCGCGCGCGCCTGCAGCAGATGGGCTTCGGAGACGTCGACCGCGACAACAAGCCCGTGTCGTTCCAACAAACGTACGAGCAGAAGCGCAGCGCCCACCTCGCCGAGATGCAGCAGAAGGAGGACGAGATGCGCCAGATGTTCGTGCAGCGGGTCAAGGAGAAGGAGGCTGAACTCAAAGAGGCCGAGAAGGAGTTGCACGCGAAGTTCGACAAGCTGAAGAAGGACCACACGGACGAGAAGAAGCGGCTGGAGGAGCTCCGCAAGAAGATCGAGGACGACGCCGTCGAGTTCAGCCGCCGCAAGCAGCTCGCCGCGCAGCCGCACCACACGCTCACCCTCGGCAAGAACAAGAAGAAGTAG
Protein
MAAIDVEPTKAEPTLRTLKLSGHVGFDSLPDQLVSKSIQNGFVFNILCIGETGLGKSTLMDSLFNTNFESSPSPHNLPTVKLKAHTYELEESNVRLKLTICDTVGYGDQVNKEDSFKAVVDYIDAQFEAYLQEELKIKRSLSSYHDSRLHVCLYFICPTGHGLKSIDLVCMKKLDTKVNIIPIIAKADTISKTELQRFKSKIMQELQSNGVEIYQFPVDDESVSEVNGSMNSHLPFAVVGSTDFVKIGNKTVRARQYPWGTVQVESESHCDFVKLREMLIRTNMEDMRDKTHARHYELYRRARLQQMGFGDVDRDNKPVSFQQTYEQKRSAHLAEMQQKEDEMRQMFVQRVKEKEAELKEAEKELHAKFDKLKKDHTDEKKRLEELRKKIEDDAVEFSRRKQLAAQPHHTLTLGKNKKK
Summary
Description
Involved in cytokinesis.
Subunit
May assemble into a multicomponent structure.
Similarity
Belongs to the TRAFAC class TrmE-Era-EngA-EngB-Septin-like GTPase superfamily. Septin GTPase family.
Keywords
Cell cycle
Cell division
Complete proteome
Cytoplasm
Cytoskeleton
GTP-binding
Mitosis
Nucleotide-binding
Reference proteome
Feature
chain Septin-2
Uniprot
H9JIN0
A0A3S2NEU5
A0A194RFJ1
A0A2A4K2N9
A0A194Q741
A0A2H1VNP7
+ More
A0A1E1WQ02 A0A212EK80 A0A2P8ZDR2 A0A087ZRQ0 E2BL51 A0A2J7R309 D2A0B5 E9IP74 E2ANZ9 A0A0K8V1H1 A0A195E5W7 A0A034WS43 A0A158N9D9 F4WJY2 A0A0A1XAG7 A0A0N0BHY7 W8C2S7 A0A154PRK3 A0A232EYT8 A0A0L7R5X9 A0A1B0DCI8 A0A195CGR9 A0A195BHT5 A0A1S4F8E6 A0A023ERV9 A0A151XIF2 A0A0A9X0K7 A0A1A9X8Y7 A0A1B0BFJ8 A0A2A3ENZ8 A0A0K8SUE2 U4UFT5 A0A182USE6 U5EUZ4 A0A195F8J4 A0A182XC30 A0A1W4XGM7 A0A1A9V164 A0A1A9X2A3 D3TMW1 A0A1B0A840 B4QS15 A0A182JVG7 A0A1Q3F2U1 A0A182PU91 A0A026WJD6 A0A182GDK8 A0A1W4VA85 B4IIA5 P54359 B0XGU0 Q29BR7 B4GN14 B4PPQ0 A0A1W4XFQ7 A0A3B0JJP5 B3M0B5 B3NYW8 A0A067RNE0 A0A1I8PMV0 B4NFV8 A0A182MY64 A0A1Y1LLT2 A0A2M4CNP8 T1PCD0 A0A182RQZ6 A0A182INV8 E0VDH1 Q7QGH0 B4JUB3 A0A084VRL7 A0A182HNC8 B4LWN0 B4K8F2 A0A182TBV5 A0A182FLM6 A0A0M4EFZ6 A0A1Q3F2Z3 A0A182Y5X2 A0A182UIC5 A0A182W2G1 A0A182LYD2 B0XKI5 A0A224XD90 A0A182QKZ0 N6TZA1 W5JSF8 A0A2M3Z6P5 A0A2M4BPJ6 A0A1L8DWP6 A0A2M4AII4 A0A2M4AII0 A0A1B0FQZ8 T1HIU7
A0A1E1WQ02 A0A212EK80 A0A2P8ZDR2 A0A087ZRQ0 E2BL51 A0A2J7R309 D2A0B5 E9IP74 E2ANZ9 A0A0K8V1H1 A0A195E5W7 A0A034WS43 A0A158N9D9 F4WJY2 A0A0A1XAG7 A0A0N0BHY7 W8C2S7 A0A154PRK3 A0A232EYT8 A0A0L7R5X9 A0A1B0DCI8 A0A195CGR9 A0A195BHT5 A0A1S4F8E6 A0A023ERV9 A0A151XIF2 A0A0A9X0K7 A0A1A9X8Y7 A0A1B0BFJ8 A0A2A3ENZ8 A0A0K8SUE2 U4UFT5 A0A182USE6 U5EUZ4 A0A195F8J4 A0A182XC30 A0A1W4XGM7 A0A1A9V164 A0A1A9X2A3 D3TMW1 A0A1B0A840 B4QS15 A0A182JVG7 A0A1Q3F2U1 A0A182PU91 A0A026WJD6 A0A182GDK8 A0A1W4VA85 B4IIA5 P54359 B0XGU0 Q29BR7 B4GN14 B4PPQ0 A0A1W4XFQ7 A0A3B0JJP5 B3M0B5 B3NYW8 A0A067RNE0 A0A1I8PMV0 B4NFV8 A0A182MY64 A0A1Y1LLT2 A0A2M4CNP8 T1PCD0 A0A182RQZ6 A0A182INV8 E0VDH1 Q7QGH0 B4JUB3 A0A084VRL7 A0A182HNC8 B4LWN0 B4K8F2 A0A182TBV5 A0A182FLM6 A0A0M4EFZ6 A0A1Q3F2Z3 A0A182Y5X2 A0A182UIC5 A0A182W2G1 A0A182LYD2 B0XKI5 A0A224XD90 A0A182QKZ0 N6TZA1 W5JSF8 A0A2M3Z6P5 A0A2M4BPJ6 A0A1L8DWP6 A0A2M4AII4 A0A2M4AII0 A0A1B0FQZ8 T1HIU7
Pubmed
19121390
26354079
22118469
29403074
20798317
18362917
+ More
19820115 21282665 25348373 21347285 21719571 25830018 24495485 28648823 24945155 25401762 26823975 23537049 20353571 17994087 24508170 26483478 10731132 12537572 12537569 15632085 17550304 24845553 28004739 25315136 20566863 12364791 24438588 25244985 20920257 23761445
19820115 21282665 25348373 21347285 21719571 25830018 24495485 28648823 24945155 25401762 26823975 23537049 20353571 17994087 24508170 26483478 10731132 12537572 12537569 15632085 17550304 24845553 28004739 25315136 20566863 12364791 24438588 25244985 20920257 23761445
EMBL
BABH01027267
RSAL01000154
RVE45675.1
KQ460436
KPJ14701.1
NWSH01000219
+ More
PCG78286.1 KQ459337 KPJ01362.1 ODYU01003521 SOQ42417.1 GDQN01005140 GDQN01002067 JAT85914.1 JAT88987.1 AGBW02014296 OWR41922.1 PYGN01000086 PSN54643.1 GL448943 EFN83569.1 NEVH01007823 PNF35227.1 KQ971338 EFA02493.1 GL764458 EFZ17616.1 GL441439 EFN64831.1 GDHF01019864 JAI32450.1 KQ979592 KYN20590.1 GAKP01001488 JAC57464.1 ADTU01009487 GL888187 EGI65504.1 GBXI01006180 JAD08112.1 KQ435737 KOX76954.1 GAMC01000313 JAC06243.1 KQ435066 KZC14367.1 NNAY01001602 OXU23460.1 KQ414648 KOC66224.1 AJVK01005126 AJVK01005127 KQ977791 KYM99919.1 KQ976476 KYM83844.1 GAPW01001658 JAC11940.1 KQ982093 KYQ60068.1 GBHO01031281 GDHC01002829 JAG12323.1 JAQ15800.1 JXJN01013502 KZ288202 PBC33533.1 GBRD01008992 JAG56829.1 KB632278 ERL91233.1 GANO01002104 JAB57767.1 KQ981727 KYN36696.1 EZ422763 ADD19039.1 CM000364 EDX12211.1 GFDL01013196 JAV21849.1 KK107214 EZA55229.1 JXUM01056137 JXUM01056138 JXUM01056139 JXUM01056140 KQ561898 KXJ77224.1 CH480842 EDW49649.1 U28966 AE014297 AY061281 DS233063 EDS27827.1 CM000070 EAL26930.1 CH479186 EDW39140.1 CM000160 EDW96150.1 OUUW01000007 SPP82574.1 CH902617 EDV43121.1 CH954181 EDV48371.1 KK852567 KDR21229.1 CH964251 EDW83175.1 GEZM01052109 JAV74602.1 GGFL01002643 MBW66821.1 KA645785 AFP60414.1 DS235078 EEB11427.1 AAAB01008834 EAA05701.3 CH916374 EDV91083.1 ATLV01015746 KE525033 KFB40611.1 APCN01001904 CH940650 EDW67695.1 CH933806 EDW16534.1 CP012526 ALC47191.1 GFDL01013121 JAV21924.1 AXCM01003888 DS233853 EDS31981.1 GFTR01006021 JAW10405.1 AXCN02002036 APGK01049283 KB741147 ENN73691.1 ADMH02000225 ETN67312.1 GGFM01003446 MBW24197.1 GGFJ01005856 MBW54997.1 GFDF01003235 JAV10849.1 GGFK01007272 MBW40593.1 GGFK01007278 MBW40599.1 CCAG010015634 CCAG010015635 ACPB03011209
PCG78286.1 KQ459337 KPJ01362.1 ODYU01003521 SOQ42417.1 GDQN01005140 GDQN01002067 JAT85914.1 JAT88987.1 AGBW02014296 OWR41922.1 PYGN01000086 PSN54643.1 GL448943 EFN83569.1 NEVH01007823 PNF35227.1 KQ971338 EFA02493.1 GL764458 EFZ17616.1 GL441439 EFN64831.1 GDHF01019864 JAI32450.1 KQ979592 KYN20590.1 GAKP01001488 JAC57464.1 ADTU01009487 GL888187 EGI65504.1 GBXI01006180 JAD08112.1 KQ435737 KOX76954.1 GAMC01000313 JAC06243.1 KQ435066 KZC14367.1 NNAY01001602 OXU23460.1 KQ414648 KOC66224.1 AJVK01005126 AJVK01005127 KQ977791 KYM99919.1 KQ976476 KYM83844.1 GAPW01001658 JAC11940.1 KQ982093 KYQ60068.1 GBHO01031281 GDHC01002829 JAG12323.1 JAQ15800.1 JXJN01013502 KZ288202 PBC33533.1 GBRD01008992 JAG56829.1 KB632278 ERL91233.1 GANO01002104 JAB57767.1 KQ981727 KYN36696.1 EZ422763 ADD19039.1 CM000364 EDX12211.1 GFDL01013196 JAV21849.1 KK107214 EZA55229.1 JXUM01056137 JXUM01056138 JXUM01056139 JXUM01056140 KQ561898 KXJ77224.1 CH480842 EDW49649.1 U28966 AE014297 AY061281 DS233063 EDS27827.1 CM000070 EAL26930.1 CH479186 EDW39140.1 CM000160 EDW96150.1 OUUW01000007 SPP82574.1 CH902617 EDV43121.1 CH954181 EDV48371.1 KK852567 KDR21229.1 CH964251 EDW83175.1 GEZM01052109 JAV74602.1 GGFL01002643 MBW66821.1 KA645785 AFP60414.1 DS235078 EEB11427.1 AAAB01008834 EAA05701.3 CH916374 EDV91083.1 ATLV01015746 KE525033 KFB40611.1 APCN01001904 CH940650 EDW67695.1 CH933806 EDW16534.1 CP012526 ALC47191.1 GFDL01013121 JAV21924.1 AXCM01003888 DS233853 EDS31981.1 GFTR01006021 JAW10405.1 AXCN02002036 APGK01049283 KB741147 ENN73691.1 ADMH02000225 ETN67312.1 GGFM01003446 MBW24197.1 GGFJ01005856 MBW54997.1 GFDF01003235 JAV10849.1 GGFK01007272 MBW40593.1 GGFK01007278 MBW40599.1 CCAG010015634 CCAG010015635 ACPB03011209
Proteomes
UP000005204
UP000283053
UP000053240
UP000218220
UP000053268
UP000007151
+ More
UP000245037 UP000005203 UP000008237 UP000235965 UP000007266 UP000000311 UP000078492 UP000005205 UP000007755 UP000053105 UP000076502 UP000215335 UP000053825 UP000092462 UP000078542 UP000078540 UP000075809 UP000092443 UP000092460 UP000242457 UP000030742 UP000075903 UP000078541 UP000076407 UP000192223 UP000078200 UP000091820 UP000092445 UP000000304 UP000075881 UP000075885 UP000053097 UP000069940 UP000249989 UP000192221 UP000001292 UP000000803 UP000002320 UP000001819 UP000008744 UP000002282 UP000268350 UP000007801 UP000008711 UP000027135 UP000095300 UP000007798 UP000075884 UP000095301 UP000075900 UP000075880 UP000009046 UP000007062 UP000001070 UP000030765 UP000075840 UP000008792 UP000009192 UP000075901 UP000069272 UP000092553 UP000076408 UP000075902 UP000075920 UP000075883 UP000075886 UP000019118 UP000000673 UP000092444 UP000015103
UP000245037 UP000005203 UP000008237 UP000235965 UP000007266 UP000000311 UP000078492 UP000005205 UP000007755 UP000053105 UP000076502 UP000215335 UP000053825 UP000092462 UP000078542 UP000078540 UP000075809 UP000092443 UP000092460 UP000242457 UP000030742 UP000075903 UP000078541 UP000076407 UP000192223 UP000078200 UP000091820 UP000092445 UP000000304 UP000075881 UP000075885 UP000053097 UP000069940 UP000249989 UP000192221 UP000001292 UP000000803 UP000002320 UP000001819 UP000008744 UP000002282 UP000268350 UP000007801 UP000008711 UP000027135 UP000095300 UP000007798 UP000075884 UP000095301 UP000075900 UP000075880 UP000009046 UP000007062 UP000001070 UP000030765 UP000075840 UP000008792 UP000009192 UP000075901 UP000069272 UP000092553 UP000076408 UP000075902 UP000075920 UP000075883 UP000075886 UP000019118 UP000000673 UP000092444 UP000015103
Pfam
PF00735 Septin
Interpro
Gene 3D
ProteinModelPortal
H9JIN0
A0A3S2NEU5
A0A194RFJ1
A0A2A4K2N9
A0A194Q741
A0A2H1VNP7
+ More
A0A1E1WQ02 A0A212EK80 A0A2P8ZDR2 A0A087ZRQ0 E2BL51 A0A2J7R309 D2A0B5 E9IP74 E2ANZ9 A0A0K8V1H1 A0A195E5W7 A0A034WS43 A0A158N9D9 F4WJY2 A0A0A1XAG7 A0A0N0BHY7 W8C2S7 A0A154PRK3 A0A232EYT8 A0A0L7R5X9 A0A1B0DCI8 A0A195CGR9 A0A195BHT5 A0A1S4F8E6 A0A023ERV9 A0A151XIF2 A0A0A9X0K7 A0A1A9X8Y7 A0A1B0BFJ8 A0A2A3ENZ8 A0A0K8SUE2 U4UFT5 A0A182USE6 U5EUZ4 A0A195F8J4 A0A182XC30 A0A1W4XGM7 A0A1A9V164 A0A1A9X2A3 D3TMW1 A0A1B0A840 B4QS15 A0A182JVG7 A0A1Q3F2U1 A0A182PU91 A0A026WJD6 A0A182GDK8 A0A1W4VA85 B4IIA5 P54359 B0XGU0 Q29BR7 B4GN14 B4PPQ0 A0A1W4XFQ7 A0A3B0JJP5 B3M0B5 B3NYW8 A0A067RNE0 A0A1I8PMV0 B4NFV8 A0A182MY64 A0A1Y1LLT2 A0A2M4CNP8 T1PCD0 A0A182RQZ6 A0A182INV8 E0VDH1 Q7QGH0 B4JUB3 A0A084VRL7 A0A182HNC8 B4LWN0 B4K8F2 A0A182TBV5 A0A182FLM6 A0A0M4EFZ6 A0A1Q3F2Z3 A0A182Y5X2 A0A182UIC5 A0A182W2G1 A0A182LYD2 B0XKI5 A0A224XD90 A0A182QKZ0 N6TZA1 W5JSF8 A0A2M3Z6P5 A0A2M4BPJ6 A0A1L8DWP6 A0A2M4AII4 A0A2M4AII0 A0A1B0FQZ8 T1HIU7
A0A1E1WQ02 A0A212EK80 A0A2P8ZDR2 A0A087ZRQ0 E2BL51 A0A2J7R309 D2A0B5 E9IP74 E2ANZ9 A0A0K8V1H1 A0A195E5W7 A0A034WS43 A0A158N9D9 F4WJY2 A0A0A1XAG7 A0A0N0BHY7 W8C2S7 A0A154PRK3 A0A232EYT8 A0A0L7R5X9 A0A1B0DCI8 A0A195CGR9 A0A195BHT5 A0A1S4F8E6 A0A023ERV9 A0A151XIF2 A0A0A9X0K7 A0A1A9X8Y7 A0A1B0BFJ8 A0A2A3ENZ8 A0A0K8SUE2 U4UFT5 A0A182USE6 U5EUZ4 A0A195F8J4 A0A182XC30 A0A1W4XGM7 A0A1A9V164 A0A1A9X2A3 D3TMW1 A0A1B0A840 B4QS15 A0A182JVG7 A0A1Q3F2U1 A0A182PU91 A0A026WJD6 A0A182GDK8 A0A1W4VA85 B4IIA5 P54359 B0XGU0 Q29BR7 B4GN14 B4PPQ0 A0A1W4XFQ7 A0A3B0JJP5 B3M0B5 B3NYW8 A0A067RNE0 A0A1I8PMV0 B4NFV8 A0A182MY64 A0A1Y1LLT2 A0A2M4CNP8 T1PCD0 A0A182RQZ6 A0A182INV8 E0VDH1 Q7QGH0 B4JUB3 A0A084VRL7 A0A182HNC8 B4LWN0 B4K8F2 A0A182TBV5 A0A182FLM6 A0A0M4EFZ6 A0A1Q3F2Z3 A0A182Y5X2 A0A182UIC5 A0A182W2G1 A0A182LYD2 B0XKI5 A0A224XD90 A0A182QKZ0 N6TZA1 W5JSF8 A0A2M3Z6P5 A0A2M4BPJ6 A0A1L8DWP6 A0A2M4AII4 A0A2M4AII0 A0A1B0FQZ8 T1HIU7
PDB
2QAG
E-value=8.78076e-152,
Score=1377
Ontologies
GO
Topology
Subcellular location
Cytoplasm
Cytoskeleton
Spindle
Cytoskeleton
Spindle
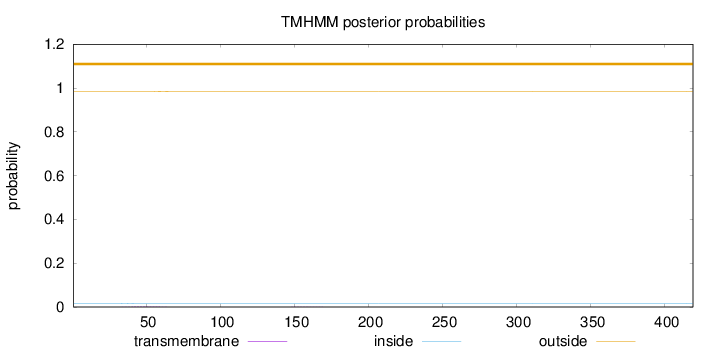
Length:
419
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0509300000000001
Exp number, first 60 AAs:
0.04267
Total prob of N-in:
0.01690
outside
1 - 419
Population Genetic Test Statistics
Pi
269.625653
Theta
128.420305
Tajima's D
-0.820819
CLR
198.127112
CSRT
0.168441577921104
Interpretation
Uncertain
