Gene
KWMTBOMO08546
Annotation
neurobeachin_[Danaus_plexippus]
Full name
Neurobeachin
Alternative Name
A-kinase anchor protein 550
Protein rugose
dAKAP550
Protein rugose
dAKAP550
Location in the cell
Mitochondrial Reliability : 2.315
Sequence
CDS
ATGTTAGCAACGTCGTCAGGCTTGAGCCCGGTACTGAAATATTGTGAGCATCTACACGGGAAATGGTACTTCAGCGAGATACGAGCTATTTTCTCGAGACGCTACCTTCTCCAGAATGTCGCCGTCGAGCTGTTCCTCGCCAGCAGAACTTCAATATTCTTCGCATTTCCCGACCAAGCGACAGTCAAGAAAGTAATCAAAGCGCTACCCCGAGTTGGTGTCGGCATCAAGTATGGCATCCCGCAGACCAGGAACAGAACTGGAGAGTTAAATAAAAGTAACAATTTAAAGAAACACTAA
Protein
MLATSSGLSPVLKYCEHLHGKWYFSEIRAIFSRRYLLQNVAVELFLASRTSIFFAFPDQATVKKVIKALPRVGVGIKYGIPQTRNRTGELNKSNNLKKH
Summary
Description
Binds to type II regulatory subunits of protein kinase A and anchors/targets them to the membrane. May anchor the kinase to cytoskeletal and/or organelle-associated proteins. Required for correct retinal pattern formation and may function in cell fate determination through its interactions with the EGFR and Notch signaling pathways. Required for associative odor learning and short-term memory. Involved in development of the neuromuscular junction and the mushroom body.
Subunit
Interacts with RII subunit of PKA and components of the EGFR-mediated and Notch-mediated signaling pathways.
Similarity
Belongs to the WD repeat neurobeachin family.
Keywords
Alternative splicing
Complete proteome
Cytoplasm
Membrane
Reference proteome
Repeat
WD repeat
Feature
chain Neurobeachin
splice variant In isoform R.
splice variant In isoform R.
Uniprot
A0A212FNE6
A0A2H1W5K8
A0A2A4J9X3
A0A194Q745
A0A0N1I6E7
B5M0U3
+ More
A0A3S2NYH0 A0A182S8U8 V9IKF6 A0A088AMU7 A0A1B6JDW0 A0A1Y1K804 A0A1Y1KFR1 A0A154PR26 A0A1Y1K827 A0A1Y1KDW4 A0A1B6GW90 A0A1B6FHD0 V5GIF1 A0A2M3YXT7 A0A1B6FFQ9 A0A1J1HWS7 D2A310 A0A232FFE5 A0A1B6GFL2 A0A1J1HSF2 A0A1W4XBX6 A0A1W4X1B0 A0A0K8TWW3 A0A1W4X1F6 A0A1W4XAS7 A0A2M4CNN2 A0A1B0BVL9 A0A1W4XAT3 A0A1W4X1B5 A0A1W4XBX0 A0A1W4X1G1 A0A1W4XC55 A0A2M4CYZ2 B0WJ16 A0A182KDJ8 A0A2M4CWG4 A0A2M4CJL6 A0A146KQW0 A0A0R1E9Z0 A0A182GPN4 A0A1B6M2V4 A0A146M1B0 A0A1I8M1Z8 A0A2M4B839 Q16FK4 A0A1I8MXG7 A0A2M4CIX4 A0A1I8M1Z5 A0A1I8M200 A0A1I8M1Z6 A0A182J3V0 A0A0A9W0E0 A0A1I8M202 A0A1I8Q9G3 A0A1I8Q9K7 A0A1W4VI88 A0A0M4EQT7 A0A1I8M203 A0A1I8Q9G7 A0A2M4CIC8 A0A1B6KZC9 A0A1B0GJH6 A0A1I8Q9H2 W5JH14 B4MJ67 A0A2M4DP17 A0A2M4CID1 B4M6T2 A0A2M4CIB8 A0A1I8Q9J9 A0A1B0G0Z8 A0A1I8Q9K4 A0A0R1EB30 B7Z0X3 A0A182X590 A0A0R1E9C2 X2JE60 A0A0R1E994 A0A1I8Q9E6 A0A182PM49 A0A453YNR8 A0A1I8Q9G8 A0A224XGR8 B4PZS6 Q9W4E2 A0A182TRD9 A0A453YNQ3 A0A182MH65 A0A182H8Q6 A0A0R1E9M6 T1HIN8 A0A3F2Z1F0 A0A1I8Q9F1 A0A1I8Q9L3
A0A3S2NYH0 A0A182S8U8 V9IKF6 A0A088AMU7 A0A1B6JDW0 A0A1Y1K804 A0A1Y1KFR1 A0A154PR26 A0A1Y1K827 A0A1Y1KDW4 A0A1B6GW90 A0A1B6FHD0 V5GIF1 A0A2M3YXT7 A0A1B6FFQ9 A0A1J1HWS7 D2A310 A0A232FFE5 A0A1B6GFL2 A0A1J1HSF2 A0A1W4XBX6 A0A1W4X1B0 A0A0K8TWW3 A0A1W4X1F6 A0A1W4XAS7 A0A2M4CNN2 A0A1B0BVL9 A0A1W4XAT3 A0A1W4X1B5 A0A1W4XBX0 A0A1W4X1G1 A0A1W4XC55 A0A2M4CYZ2 B0WJ16 A0A182KDJ8 A0A2M4CWG4 A0A2M4CJL6 A0A146KQW0 A0A0R1E9Z0 A0A182GPN4 A0A1B6M2V4 A0A146M1B0 A0A1I8M1Z8 A0A2M4B839 Q16FK4 A0A1I8MXG7 A0A2M4CIX4 A0A1I8M1Z5 A0A1I8M200 A0A1I8M1Z6 A0A182J3V0 A0A0A9W0E0 A0A1I8M202 A0A1I8Q9G3 A0A1I8Q9K7 A0A1W4VI88 A0A0M4EQT7 A0A1I8M203 A0A1I8Q9G7 A0A2M4CIC8 A0A1B6KZC9 A0A1B0GJH6 A0A1I8Q9H2 W5JH14 B4MJ67 A0A2M4DP17 A0A2M4CID1 B4M6T2 A0A2M4CIB8 A0A1I8Q9J9 A0A1B0G0Z8 A0A1I8Q9K4 A0A0R1EB30 B7Z0X3 A0A182X590 A0A0R1E9C2 X2JE60 A0A0R1E994 A0A1I8Q9E6 A0A182PM49 A0A453YNR8 A0A1I8Q9G8 A0A224XGR8 B4PZS6 Q9W4E2 A0A182TRD9 A0A453YNQ3 A0A182MH65 A0A182H8Q6 A0A0R1E9M6 T1HIN8 A0A3F2Z1F0 A0A1I8Q9F1 A0A1I8Q9L3
Pubmed
EMBL
AGBW02006209
OWR55229.1
ODYU01006172
SOQ47794.1
NWSH01002199
PCG68927.1
+ More
KQ459337 KPJ01367.1 KQ460817 KPJ12121.1 EU930279 ACH56907.1 RSAL01000024 RVE52214.1 JR049953 AEY61137.1 GECU01010389 JAS97317.1 GEZM01089253 GEZM01089252 JAV57592.1 GEZM01089254 GEZM01089249 JAV57597.1 KQ435066 KZC14375.1 GEZM01089251 GEZM01089248 JAV57599.1 GEZM01089250 JAV57596.1 GECZ01003156 JAS66613.1 GECZ01020160 JAS49609.1 GALX01004627 JAB63839.1 GGFM01000341 MBW21092.1 GECZ01020723 JAS49046.1 CVRI01000020 CRK90982.1 KQ971338 EFA01972.2 NNAY01000293 OXU29415.1 GECZ01008527 JAS61242.1 CRK90983.1 GDHF01033362 JAI18952.1 GGFL01002725 MBW66903.1 JXJN01021367 JXJN01021368 GGFL01005900 MBW70078.1 DS231955 EDS28895.1 GGFL01005431 MBW69609.1 GGFL01001257 MBW65435.1 GDHC01021007 JAP97621.1 CM000162 KRK05983.1 JXUM01078790 KQ563088 KXJ74501.1 GEBQ01009747 JAT30230.1 GDHC01005584 JAQ13045.1 GGFJ01000019 MBW49160.1 CH478422 EAT33014.1 GGFL01001027 MBW65205.1 GBHO01041707 JAG01897.1 CP012528 ALC48352.1 GGFL01000919 MBW65097.1 GEBQ01023170 JAT16807.1 AJWK01021725 AJWK01021726 AJWK01021727 AJWK01021728 AJWK01021729 AJWK01021730 AJWK01021731 AJWK01021732 AJWK01021733 AJWK01021734 ADMH02001415 ETN62613.1 CH963738 EDW72156.2 GGFL01015148 MBW79326.1 GGFL01000916 MBW65094.1 CH940653 EDW62499.2 GGFL01000918 MBW65096.1 CCAG010000760 CCAG010000761 CCAG010000762 CCAG010000763 CCAG010000764 KRK05982.1 AE014298 ACL82890.2 KRK05980.1 AHN59349.1 KRK05984.1 AAAB01008846 GFTR01008776 JAW07650.1 EDX01143.2 Y18278 AF003622 AY051596 AXCM01002696 AXCM01002697 JXUM01029235 JXUM01029236 JXUM01029237 JXUM01029238 JXUM01029239 JXUM01029240 JXUM01029241 JXUM01029242 JXUM01029243 JXUM01029244 KRK05981.1 ACPB03011228 ACPB03011229 ACPB03011230
KQ459337 KPJ01367.1 KQ460817 KPJ12121.1 EU930279 ACH56907.1 RSAL01000024 RVE52214.1 JR049953 AEY61137.1 GECU01010389 JAS97317.1 GEZM01089253 GEZM01089252 JAV57592.1 GEZM01089254 GEZM01089249 JAV57597.1 KQ435066 KZC14375.1 GEZM01089251 GEZM01089248 JAV57599.1 GEZM01089250 JAV57596.1 GECZ01003156 JAS66613.1 GECZ01020160 JAS49609.1 GALX01004627 JAB63839.1 GGFM01000341 MBW21092.1 GECZ01020723 JAS49046.1 CVRI01000020 CRK90982.1 KQ971338 EFA01972.2 NNAY01000293 OXU29415.1 GECZ01008527 JAS61242.1 CRK90983.1 GDHF01033362 JAI18952.1 GGFL01002725 MBW66903.1 JXJN01021367 JXJN01021368 GGFL01005900 MBW70078.1 DS231955 EDS28895.1 GGFL01005431 MBW69609.1 GGFL01001257 MBW65435.1 GDHC01021007 JAP97621.1 CM000162 KRK05983.1 JXUM01078790 KQ563088 KXJ74501.1 GEBQ01009747 JAT30230.1 GDHC01005584 JAQ13045.1 GGFJ01000019 MBW49160.1 CH478422 EAT33014.1 GGFL01001027 MBW65205.1 GBHO01041707 JAG01897.1 CP012528 ALC48352.1 GGFL01000919 MBW65097.1 GEBQ01023170 JAT16807.1 AJWK01021725 AJWK01021726 AJWK01021727 AJWK01021728 AJWK01021729 AJWK01021730 AJWK01021731 AJWK01021732 AJWK01021733 AJWK01021734 ADMH02001415 ETN62613.1 CH963738 EDW72156.2 GGFL01015148 MBW79326.1 GGFL01000916 MBW65094.1 CH940653 EDW62499.2 GGFL01000918 MBW65096.1 CCAG010000760 CCAG010000761 CCAG010000762 CCAG010000763 CCAG010000764 KRK05982.1 AE014298 ACL82890.2 KRK05980.1 AHN59349.1 KRK05984.1 AAAB01008846 GFTR01008776 JAW07650.1 EDX01143.2 Y18278 AF003622 AY051596 AXCM01002696 AXCM01002697 JXUM01029235 JXUM01029236 JXUM01029237 JXUM01029238 JXUM01029239 JXUM01029240 JXUM01029241 JXUM01029242 JXUM01029243 JXUM01029244 KRK05981.1 ACPB03011228 ACPB03011229 ACPB03011230
Proteomes
UP000007151
UP000218220
UP000053268
UP000053240
UP000283053
UP000075901
+ More
UP000005203 UP000076502 UP000183832 UP000007266 UP000215335 UP000192223 UP000092460 UP000002320 UP000075881 UP000002282 UP000069940 UP000249989 UP000095301 UP000008820 UP000075880 UP000095300 UP000192221 UP000092553 UP000092461 UP000000673 UP000007798 UP000008792 UP000092444 UP000000803 UP000076407 UP000075885 UP000075902 UP000075883 UP000015103
UP000005203 UP000076502 UP000183832 UP000007266 UP000215335 UP000192223 UP000092460 UP000002320 UP000075881 UP000002282 UP000069940 UP000249989 UP000095301 UP000008820 UP000075880 UP000095300 UP000192221 UP000092553 UP000092461 UP000000673 UP000007798 UP000008792 UP000092444 UP000000803 UP000076407 UP000075885 UP000075902 UP000075883 UP000015103
PRIDE
Pfam
Interpro
IPR023362
PH-BEACH_dom
+ More
IPR000409 BEACH_dom
IPR036372 BEACH_dom_sf
IPR036322 WD40_repeat_dom_sf
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR010508 DUF1088
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR016024 ARM-type_fold
IPR031570 DUF4704
IPR013320 ConA-like_dom_sf
IPR000253 FHA_dom
IPR022613 CAMSAP_CH
IPR031372 CAMSAP_CC1
IPR001715 CH-domain
IPR036872 CH_dom_sf
IPR000409 BEACH_dom
IPR036372 BEACH_dom_sf
IPR036322 WD40_repeat_dom_sf
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR010508 DUF1088
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR016024 ARM-type_fold
IPR031570 DUF4704
IPR013320 ConA-like_dom_sf
IPR000253 FHA_dom
IPR022613 CAMSAP_CH
IPR031372 CAMSAP_CC1
IPR001715 CH-domain
IPR036872 CH_dom_sf
SUPFAM
Gene 3D
ProteinModelPortal
A0A212FNE6
A0A2H1W5K8
A0A2A4J9X3
A0A194Q745
A0A0N1I6E7
B5M0U3
+ More
A0A3S2NYH0 A0A182S8U8 V9IKF6 A0A088AMU7 A0A1B6JDW0 A0A1Y1K804 A0A1Y1KFR1 A0A154PR26 A0A1Y1K827 A0A1Y1KDW4 A0A1B6GW90 A0A1B6FHD0 V5GIF1 A0A2M3YXT7 A0A1B6FFQ9 A0A1J1HWS7 D2A310 A0A232FFE5 A0A1B6GFL2 A0A1J1HSF2 A0A1W4XBX6 A0A1W4X1B0 A0A0K8TWW3 A0A1W4X1F6 A0A1W4XAS7 A0A2M4CNN2 A0A1B0BVL9 A0A1W4XAT3 A0A1W4X1B5 A0A1W4XBX0 A0A1W4X1G1 A0A1W4XC55 A0A2M4CYZ2 B0WJ16 A0A182KDJ8 A0A2M4CWG4 A0A2M4CJL6 A0A146KQW0 A0A0R1E9Z0 A0A182GPN4 A0A1B6M2V4 A0A146M1B0 A0A1I8M1Z8 A0A2M4B839 Q16FK4 A0A1I8MXG7 A0A2M4CIX4 A0A1I8M1Z5 A0A1I8M200 A0A1I8M1Z6 A0A182J3V0 A0A0A9W0E0 A0A1I8M202 A0A1I8Q9G3 A0A1I8Q9K7 A0A1W4VI88 A0A0M4EQT7 A0A1I8M203 A0A1I8Q9G7 A0A2M4CIC8 A0A1B6KZC9 A0A1B0GJH6 A0A1I8Q9H2 W5JH14 B4MJ67 A0A2M4DP17 A0A2M4CID1 B4M6T2 A0A2M4CIB8 A0A1I8Q9J9 A0A1B0G0Z8 A0A1I8Q9K4 A0A0R1EB30 B7Z0X3 A0A182X590 A0A0R1E9C2 X2JE60 A0A0R1E994 A0A1I8Q9E6 A0A182PM49 A0A453YNR8 A0A1I8Q9G8 A0A224XGR8 B4PZS6 Q9W4E2 A0A182TRD9 A0A453YNQ3 A0A182MH65 A0A182H8Q6 A0A0R1E9M6 T1HIN8 A0A3F2Z1F0 A0A1I8Q9F1 A0A1I8Q9L3
A0A3S2NYH0 A0A182S8U8 V9IKF6 A0A088AMU7 A0A1B6JDW0 A0A1Y1K804 A0A1Y1KFR1 A0A154PR26 A0A1Y1K827 A0A1Y1KDW4 A0A1B6GW90 A0A1B6FHD0 V5GIF1 A0A2M3YXT7 A0A1B6FFQ9 A0A1J1HWS7 D2A310 A0A232FFE5 A0A1B6GFL2 A0A1J1HSF2 A0A1W4XBX6 A0A1W4X1B0 A0A0K8TWW3 A0A1W4X1F6 A0A1W4XAS7 A0A2M4CNN2 A0A1B0BVL9 A0A1W4XAT3 A0A1W4X1B5 A0A1W4XBX0 A0A1W4X1G1 A0A1W4XC55 A0A2M4CYZ2 B0WJ16 A0A182KDJ8 A0A2M4CWG4 A0A2M4CJL6 A0A146KQW0 A0A0R1E9Z0 A0A182GPN4 A0A1B6M2V4 A0A146M1B0 A0A1I8M1Z8 A0A2M4B839 Q16FK4 A0A1I8MXG7 A0A2M4CIX4 A0A1I8M1Z5 A0A1I8M200 A0A1I8M1Z6 A0A182J3V0 A0A0A9W0E0 A0A1I8M202 A0A1I8Q9G3 A0A1I8Q9K7 A0A1W4VI88 A0A0M4EQT7 A0A1I8M203 A0A1I8Q9G7 A0A2M4CIC8 A0A1B6KZC9 A0A1B0GJH6 A0A1I8Q9H2 W5JH14 B4MJ67 A0A2M4DP17 A0A2M4CID1 B4M6T2 A0A2M4CIB8 A0A1I8Q9J9 A0A1B0G0Z8 A0A1I8Q9K4 A0A0R1EB30 B7Z0X3 A0A182X590 A0A0R1E9C2 X2JE60 A0A0R1E994 A0A1I8Q9E6 A0A182PM49 A0A453YNR8 A0A1I8Q9G8 A0A224XGR8 B4PZS6 Q9W4E2 A0A182TRD9 A0A453YNQ3 A0A182MH65 A0A182H8Q6 A0A0R1E9M6 T1HIN8 A0A3F2Z1F0 A0A1I8Q9F1 A0A1I8Q9L3
PDB
1MI1
E-value=2.12924e-28,
Score=306
Ontologies
GO
Topology
Subcellular location
Cytoplasm
Membrane
Perikaryon
Membrane
Perikaryon
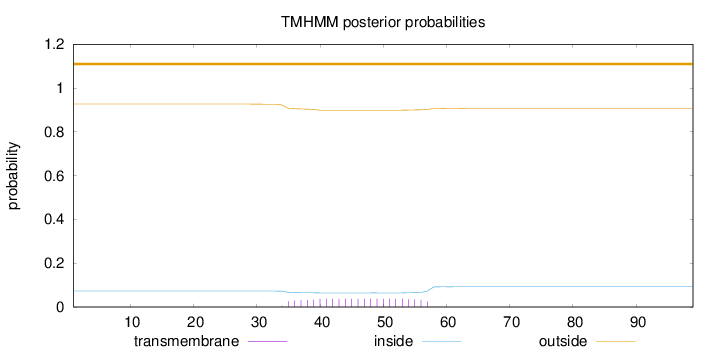
Length:
99
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.81342
Exp number, first 60 AAs:
0.81138
Total prob of N-in:
0.07324
outside
1 - 99
Population Genetic Test Statistics
Pi
195.066226
Theta
153.446468
Tajima's D
0.619573
CLR
1.219875
CSRT
0.551722413879306
Interpretation
Uncertain
