Gene
KWMTBOMO08545
Pre Gene Modal
BGIBMGA009376
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_facilitated_trehalose_transporter_Tret1-2_homolog_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.981
Sequence
CDS
ATGAGTATTGGACTCCCTACATCAGTTGCGGGGGCGTTAATGCTGAGAATCGTTGTGGGGGTGTTGGCGGCCCTTGCCGCGCAATCTATCAACATATCGGTGGGTTTCTGTCAGGGCTTCTCGGCGGTTCTGCTGCCCCAGTACTCGCTTGAGTATCCGTCCATATCGTACGAACAGAGCTCATGGATTGCAAGTCTGGGTGTGATCTCCAACCCCATTGGAGCGCTGCTGGGCGGCATGATGGTGGACGCAGTGGGGCGGCGGCTGCTGTTGCAGTCGATCGTGCTGCCGAACCTCATAGGCTGGCTGGTCATAGCCTTCTCAGAGACCTACGTCTTCCTCTGCATAGGACGATTCATCACCGGATTCACTATCGGAATGTCAACGGTATCGTACATCTACGTGGCTGAGATCACGACTCCAGAAAAGAGAGGGGTTCTCAGCGCTTTAGGGCCCGGCCTGGTCTCCACCGGGATATTCATCGTCTACTCCTTAGGAGCCTTCATTCACTGGCGAAAAGTGGCCGCTATTTGTGCGGGAGTGTCTCTACTGACTCCCTTCTTGATGTACTTCGTTCCAGAGTCACCTCTGTGGCTCGCCTCTAAAGGACAAATGAAAGAAGCTTACAACGCTATGTTCTGGCTCCGTCAGAACAACAGCACGGCGCAACAAGAACTCATGGAGTTCACCAAAGACCGCAAGGAACAGCAGATGAACTTCAAACAGAAATTAGCACTCTTCAAACGGAAAAACGTCCTCAAGCCCTTCGTATTATTAATAGTTTTCTTCATCTTCCAAGAAATGTCAGGGATTTACATCATATTGTATTACGCTGTGGATTTCTTCAAGTCTGTAGGTACTAGCGTCAACGAGTTCACGGCTTCCATCATAGTGGGAGGTGTGAGAGTCTTCATGGGAGCAGTCGGCGCTTGTCTTATTAACAGCTTCAGAAGGAAGACCCTGGCAGCAGTATCAGGACTGCTGCTGGGCATCGCCATGCTGGGGGCCGCGGTGTGCGACAGTCTCAATGGACCTCCGCTAGTGAAGCTCGGCTGTGTGCTATTACACGTTTCCTTCAGCATGGTCGGCTTCCTCCAACTGCCCTGGATCATGTCCGGAGAACTGTACCCTCAGGATATCAGAGGAGTCATGTCCGGGGCGACTTCGTGCTGCGCTTACGTCCTCATTTTCTTCAACATAAAAACGTATCCTAAATTGGAGAACTTACTCACGAGCAACGGGACTTTGTATTTGTTCGGGTTCTGTGCAATGATAGGAGCGGCATACTGCTTCCTGTTTCTGCCGGAGACTAAAGGGAAGACTCTCACGGAAATCATGAGGCAGTTCGACGATGATAACAAAAATGAGAAAGATCTTGAAGCCGGCTACAAGAGCGATAAGGGATGCAGTATCGAGGGCAGGCCAGTGCAGAGGAGACACAGCGCTGGAGCCGCTGTTTCTTTAGAAAAGAGCGAAGACTTTAAGCTTTGGAAGCCAGAAGAGTCGAATAAAACGGACACTCATTGTGATTGA
Protein
MSIGLPTSVAGALMLRIVVGVLAALAAQSINISVGFCQGFSAVLLPQYSLEYPSISYEQSSWIASLGVISNPIGALLGGMMVDAVGRRLLLQSIVLPNLIGWLVIAFSETYVFLCIGRFITGFTIGMSTVSYIYVAEITTPEKRGVLSALGPGLVSTGIFIVYSLGAFIHWRKVAAICAGVSLLTPFLMYFVPESPLWLASKGQMKEAYNAMFWLRQNNSTAQQELMEFTKDRKEQQMNFKQKLALFKRKNVLKPFVLLIVFFIFQEMSGIYIILYYAVDFFKSVGTSVNEFTASIIVGGVRVFMGAVGACLINSFRRKTLAAVSGLLLGIAMLGAAVCDSLNGPPLVKLGCVLLHVSFSMVGFLQLPWIMSGELYPQDIRGVMSGATSCCAYVLIFFNIKTYPKLENLLTSNGTLYLFGFCAMIGAAYCFLFLPETKGKTLTEIMRQFDDDNKNEKDLEAGYKSDKGCSIEGRPVQRRHSAGAAVSLEKSEDFKLWKPEESNKTDTHCD
Summary
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Uniprot
A0A2H1W435
A0A212FNG2
A0A2A4JB59
H9JIM6
A0A0N0PBS7
A0A194Q762
+ More
A0A0L7LMY4 A0A067R1A9 A0A2J7PH27 A0A2A3E7T1 A0A158P3D9 A0A195DNA9 E2BQ52 A0A1B6KFT6 A0A1B6M773 J9JJ40 E9IPE9 A0A2S2NAW7 A0A087ZSF6 A0A2H8TJ96 A0A146LNZ1 A0A0C9RG22 A0A0A9X982 A0A0A8J8M8 T1I6V7 A0A0J7L5D2 A0A3L8DR99 A0A310SL43 A0A1B6DNR0 A0A2P8YWI6 A0A0L7RCT5 D2A4W4 A0A154P3W1 A0A1B6BY83 E2AUM5 A0A2R7VTA9 A0A3Q0J6J4 E0VSU5 T1IBF1 A0A0A8J7H4 A0A1Q3FZS3 A0A1B6LM35 A0A1B6LRS5 A0A1B6FBR5 A0A1Q3FZR1 A0A084WU29 A0A2S2QED2 A0A069DUH3 A0A1B6J8E0 Q7Q1K3 A0A182IKM6 A0A1S4H1Y8 A0A023F0Y2 A0A182F0X3 A0A2M4BK99 A0A0V0G9E5 T1I7D1 A0A182V1G2 A0A182KU30 A0A182HRX4 A0A182XN87 A0A2M4CS35 A0A2C9GLD0 A0A182M3T9 Q16TA1 A0A182Y0N5 A0A182NBR2 A0A182JYR6 A0A182WGI8 A0A182T850 A0A182PZC7 A0A182TTM5 Q7QJU9 A0A182I3T0 A0A182WIV2 A0A182PQ17 A0A182FCR5 A0A182X6H8 A0A182KRX2 A0A182UPZ6 A0A026VZD3 A0A182R3T4 A0A154P1X7 A0A182GVD4 A0A3L8DT23 A0A0T6AWG8 A0A2J7RF80 A0A3Q0INK6 A0A2R7VRY9 E9ICR4 A0A1S4EC87 A0A1Y0AWP3 A0A182J2R0 A0A1Y0AWP4
A0A0L7LMY4 A0A067R1A9 A0A2J7PH27 A0A2A3E7T1 A0A158P3D9 A0A195DNA9 E2BQ52 A0A1B6KFT6 A0A1B6M773 J9JJ40 E9IPE9 A0A2S2NAW7 A0A087ZSF6 A0A2H8TJ96 A0A146LNZ1 A0A0C9RG22 A0A0A9X982 A0A0A8J8M8 T1I6V7 A0A0J7L5D2 A0A3L8DR99 A0A310SL43 A0A1B6DNR0 A0A2P8YWI6 A0A0L7RCT5 D2A4W4 A0A154P3W1 A0A1B6BY83 E2AUM5 A0A2R7VTA9 A0A3Q0J6J4 E0VSU5 T1IBF1 A0A0A8J7H4 A0A1Q3FZS3 A0A1B6LM35 A0A1B6LRS5 A0A1B6FBR5 A0A1Q3FZR1 A0A084WU29 A0A2S2QED2 A0A069DUH3 A0A1B6J8E0 Q7Q1K3 A0A182IKM6 A0A1S4H1Y8 A0A023F0Y2 A0A182F0X3 A0A2M4BK99 A0A0V0G9E5 T1I7D1 A0A182V1G2 A0A182KU30 A0A182HRX4 A0A182XN87 A0A2M4CS35 A0A2C9GLD0 A0A182M3T9 Q16TA1 A0A182Y0N5 A0A182NBR2 A0A182JYR6 A0A182WGI8 A0A182T850 A0A182PZC7 A0A182TTM5 Q7QJU9 A0A182I3T0 A0A182WIV2 A0A182PQ17 A0A182FCR5 A0A182X6H8 A0A182KRX2 A0A182UPZ6 A0A026VZD3 A0A182R3T4 A0A154P1X7 A0A182GVD4 A0A3L8DT23 A0A0T6AWG8 A0A2J7RF80 A0A3Q0INK6 A0A2R7VRY9 E9ICR4 A0A1S4EC87 A0A1Y0AWP3 A0A182J2R0 A0A1Y0AWP4
Pubmed
EMBL
ODYU01006172
SOQ47793.1
AGBW02006209
OWR55230.1
NWSH01002199
PCG68928.1
+ More
BABH01027231 KQ460817 KPJ12122.1 KQ459337 KPJ01368.1 JTDY01000523 KOB76807.1 KK852817 KDR15737.1 NEVH01025154 PNF15629.1 KZ288340 PBC27750.1 ADTU01007656 KQ980713 KYN14358.1 GL449698 EFN82171.1 GEBQ01029692 JAT10285.1 GEBQ01008190 JAT31787.1 ABLF02018355 GL764503 EFZ17558.1 GGMR01001742 MBY14361.1 GFXV01002380 MBW14185.1 GDHC01010282 GDHC01003621 JAQ08347.1 JAQ15008.1 GBYB01007274 GBYB01007275 JAG77041.1 JAG77042.1 GBHO01028221 JAG15383.1 AB901064 BAQ02364.1 ACPB03010141 LBMM01000704 KMQ97694.1 QOIP01000005 RLU22921.1 KQ764883 OAD54259.1 GEDC01017793 GEDC01009990 JAS19505.1 JAS27308.1 PYGN01000318 PSN48610.1 KQ414615 KOC68663.1 KQ971344 EFA05276.2 KQ434796 KZC05820.1 GEDC01031065 GEDC01025950 JAS06233.1 JAS11348.1 GL442829 EFN62872.1 KK854065 PTY10579.1 DS235758 EEB16451.1 ACPB03005679 AB901070 BAQ02370.1 GFDL01001987 JAV33058.1 GEBQ01015348 JAT24629.1 GEBQ01013595 JAT26382.1 GECZ01022112 JAS47657.1 GFDL01001990 JAV33055.1 ATLV01026995 KE525421 KFB53723.1 GGMS01006902 MBY76105.1 GBGD01001284 JAC87605.1 GECU01012268 JAS95438.1 AAAB01008980 EAA14209.4 GBBI01004041 JAC14671.1 GGFJ01004336 MBW53477.1 GECL01001732 JAP04392.1 ACPB03015438 ACPB03015439 APCN01001774 GGFL01003976 MBW68154.1 AXCM01001091 CH477657 EAT37688.1 AXCN02000684 AAAB01008807 EAA04222.4 APCN01000220 KK107538 EZA49138.1 KZC05843.1 JXUM01090735 KQ563861 KXJ73152.1 RLU22918.1 LJIG01022656 KRT79410.1 NEVH01004413 PNF39494.1 KK854045 PTY10206.1 GL762354 EFZ21649.1 KY921826 ART29415.1 KY921829 ART29418.1
BABH01027231 KQ460817 KPJ12122.1 KQ459337 KPJ01368.1 JTDY01000523 KOB76807.1 KK852817 KDR15737.1 NEVH01025154 PNF15629.1 KZ288340 PBC27750.1 ADTU01007656 KQ980713 KYN14358.1 GL449698 EFN82171.1 GEBQ01029692 JAT10285.1 GEBQ01008190 JAT31787.1 ABLF02018355 GL764503 EFZ17558.1 GGMR01001742 MBY14361.1 GFXV01002380 MBW14185.1 GDHC01010282 GDHC01003621 JAQ08347.1 JAQ15008.1 GBYB01007274 GBYB01007275 JAG77041.1 JAG77042.1 GBHO01028221 JAG15383.1 AB901064 BAQ02364.1 ACPB03010141 LBMM01000704 KMQ97694.1 QOIP01000005 RLU22921.1 KQ764883 OAD54259.1 GEDC01017793 GEDC01009990 JAS19505.1 JAS27308.1 PYGN01000318 PSN48610.1 KQ414615 KOC68663.1 KQ971344 EFA05276.2 KQ434796 KZC05820.1 GEDC01031065 GEDC01025950 JAS06233.1 JAS11348.1 GL442829 EFN62872.1 KK854065 PTY10579.1 DS235758 EEB16451.1 ACPB03005679 AB901070 BAQ02370.1 GFDL01001987 JAV33058.1 GEBQ01015348 JAT24629.1 GEBQ01013595 JAT26382.1 GECZ01022112 JAS47657.1 GFDL01001990 JAV33055.1 ATLV01026995 KE525421 KFB53723.1 GGMS01006902 MBY76105.1 GBGD01001284 JAC87605.1 GECU01012268 JAS95438.1 AAAB01008980 EAA14209.4 GBBI01004041 JAC14671.1 GGFJ01004336 MBW53477.1 GECL01001732 JAP04392.1 ACPB03015438 ACPB03015439 APCN01001774 GGFL01003976 MBW68154.1 AXCM01001091 CH477657 EAT37688.1 AXCN02000684 AAAB01008807 EAA04222.4 APCN01000220 KK107538 EZA49138.1 KZC05843.1 JXUM01090735 KQ563861 KXJ73152.1 RLU22918.1 LJIG01022656 KRT79410.1 NEVH01004413 PNF39494.1 KK854045 PTY10206.1 GL762354 EFZ21649.1 KY921826 ART29415.1 KY921829 ART29418.1
Proteomes
UP000007151
UP000218220
UP000005204
UP000053240
UP000053268
UP000037510
+ More
UP000027135 UP000235965 UP000242457 UP000005205 UP000078492 UP000008237 UP000007819 UP000005203 UP000015103 UP000036403 UP000279307 UP000245037 UP000053825 UP000007266 UP000076502 UP000000311 UP000079169 UP000009046 UP000030765 UP000007062 UP000075880 UP000069272 UP000075903 UP000075882 UP000075840 UP000076407 UP000075883 UP000008820 UP000076408 UP000075884 UP000075881 UP000075920 UP000075901 UP000075886 UP000075902 UP000075885 UP000053097 UP000075900 UP000069940 UP000249989
UP000027135 UP000235965 UP000242457 UP000005205 UP000078492 UP000008237 UP000007819 UP000005203 UP000015103 UP000036403 UP000279307 UP000245037 UP000053825 UP000007266 UP000076502 UP000000311 UP000079169 UP000009046 UP000030765 UP000007062 UP000075880 UP000069272 UP000075903 UP000075882 UP000075840 UP000076407 UP000075883 UP000008820 UP000076408 UP000075884 UP000075881 UP000075920 UP000075901 UP000075886 UP000075902 UP000075885 UP000053097 UP000075900 UP000069940 UP000249989
PRIDE
Pfam
PF00083 Sugar_tr
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
A0A2H1W435
A0A212FNG2
A0A2A4JB59
H9JIM6
A0A0N0PBS7
A0A194Q762
+ More
A0A0L7LMY4 A0A067R1A9 A0A2J7PH27 A0A2A3E7T1 A0A158P3D9 A0A195DNA9 E2BQ52 A0A1B6KFT6 A0A1B6M773 J9JJ40 E9IPE9 A0A2S2NAW7 A0A087ZSF6 A0A2H8TJ96 A0A146LNZ1 A0A0C9RG22 A0A0A9X982 A0A0A8J8M8 T1I6V7 A0A0J7L5D2 A0A3L8DR99 A0A310SL43 A0A1B6DNR0 A0A2P8YWI6 A0A0L7RCT5 D2A4W4 A0A154P3W1 A0A1B6BY83 E2AUM5 A0A2R7VTA9 A0A3Q0J6J4 E0VSU5 T1IBF1 A0A0A8J7H4 A0A1Q3FZS3 A0A1B6LM35 A0A1B6LRS5 A0A1B6FBR5 A0A1Q3FZR1 A0A084WU29 A0A2S2QED2 A0A069DUH3 A0A1B6J8E0 Q7Q1K3 A0A182IKM6 A0A1S4H1Y8 A0A023F0Y2 A0A182F0X3 A0A2M4BK99 A0A0V0G9E5 T1I7D1 A0A182V1G2 A0A182KU30 A0A182HRX4 A0A182XN87 A0A2M4CS35 A0A2C9GLD0 A0A182M3T9 Q16TA1 A0A182Y0N5 A0A182NBR2 A0A182JYR6 A0A182WGI8 A0A182T850 A0A182PZC7 A0A182TTM5 Q7QJU9 A0A182I3T0 A0A182WIV2 A0A182PQ17 A0A182FCR5 A0A182X6H8 A0A182KRX2 A0A182UPZ6 A0A026VZD3 A0A182R3T4 A0A154P1X7 A0A182GVD4 A0A3L8DT23 A0A0T6AWG8 A0A2J7RF80 A0A3Q0INK6 A0A2R7VRY9 E9ICR4 A0A1S4EC87 A0A1Y0AWP3 A0A182J2R0 A0A1Y0AWP4
A0A0L7LMY4 A0A067R1A9 A0A2J7PH27 A0A2A3E7T1 A0A158P3D9 A0A195DNA9 E2BQ52 A0A1B6KFT6 A0A1B6M773 J9JJ40 E9IPE9 A0A2S2NAW7 A0A087ZSF6 A0A2H8TJ96 A0A146LNZ1 A0A0C9RG22 A0A0A9X982 A0A0A8J8M8 T1I6V7 A0A0J7L5D2 A0A3L8DR99 A0A310SL43 A0A1B6DNR0 A0A2P8YWI6 A0A0L7RCT5 D2A4W4 A0A154P3W1 A0A1B6BY83 E2AUM5 A0A2R7VTA9 A0A3Q0J6J4 E0VSU5 T1IBF1 A0A0A8J7H4 A0A1Q3FZS3 A0A1B6LM35 A0A1B6LRS5 A0A1B6FBR5 A0A1Q3FZR1 A0A084WU29 A0A2S2QED2 A0A069DUH3 A0A1B6J8E0 Q7Q1K3 A0A182IKM6 A0A1S4H1Y8 A0A023F0Y2 A0A182F0X3 A0A2M4BK99 A0A0V0G9E5 T1I7D1 A0A182V1G2 A0A182KU30 A0A182HRX4 A0A182XN87 A0A2M4CS35 A0A2C9GLD0 A0A182M3T9 Q16TA1 A0A182Y0N5 A0A182NBR2 A0A182JYR6 A0A182WGI8 A0A182T850 A0A182PZC7 A0A182TTM5 Q7QJU9 A0A182I3T0 A0A182WIV2 A0A182PQ17 A0A182FCR5 A0A182X6H8 A0A182KRX2 A0A182UPZ6 A0A026VZD3 A0A182R3T4 A0A154P1X7 A0A182GVD4 A0A3L8DT23 A0A0T6AWG8 A0A2J7RF80 A0A3Q0INK6 A0A2R7VRY9 E9ICR4 A0A1S4EC87 A0A1Y0AWP3 A0A182J2R0 A0A1Y0AWP4
PDB
4LDS
E-value=1.44872e-27,
Score=307
Ontologies
GO
Topology
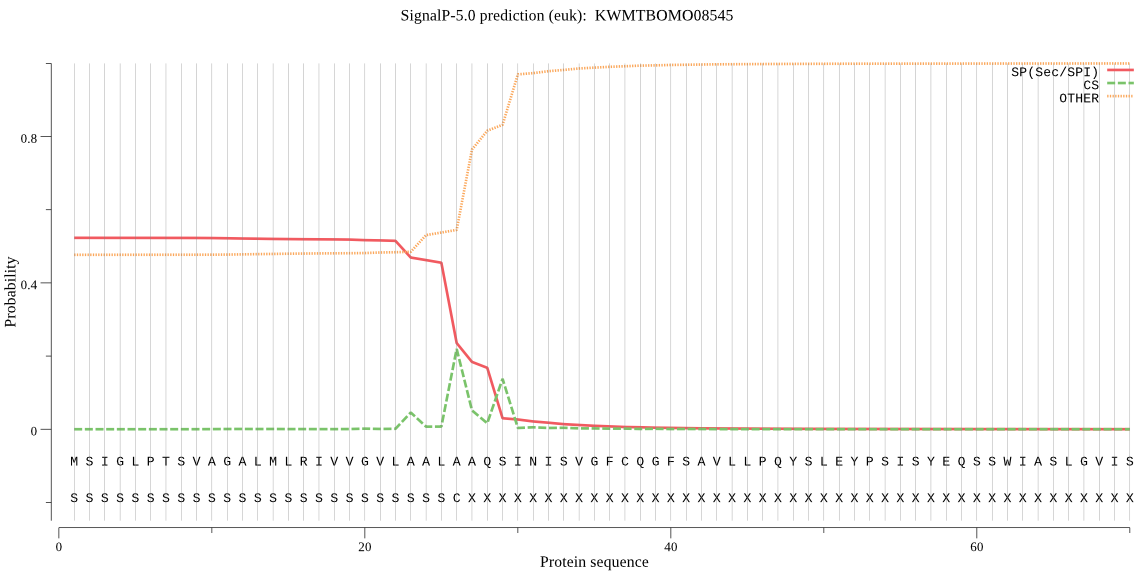
SignalP
Position: 1 - 26,
Likelihood: 0.523206
Length:
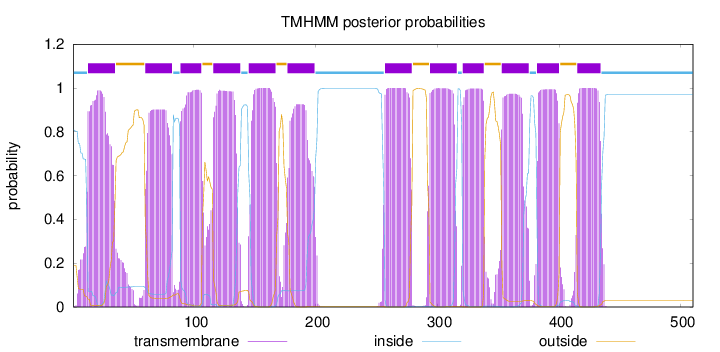
510
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
253.68615
Exp number, first 60 AAs:
25.32108
Total prob of N-in:
0.80945
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 35
outside
36 - 59
TMhelix
60 - 82
inside
83 - 88
TMhelix
89 - 106
outside
107 - 115
TMhelix
116 - 138
inside
139 - 144
TMhelix
145 - 167
outside
168 - 176
TMhelix
177 - 199
inside
200 - 256
TMhelix
257 - 279
outside
280 - 293
TMhelix
294 - 316
inside
317 - 320
TMhelix
321 - 338
outside
339 - 352
TMhelix
353 - 375
inside
376 - 381
TMhelix
382 - 400
outside
401 - 414
TMhelix
415 - 434
inside
435 - 510
Population Genetic Test Statistics
Pi
229.012406
Theta
195.219863
Tajima's D
0.512081
CLR
0.352705
CSRT
0.523473826308685
Interpretation
Uncertain
