Gene
KWMTBOMO08544
Pre Gene Modal
BGIBMGA009317
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_neurobeachin-like_[Bombyx_mori]
Full name
Neurobeachin
Alternative Name
A-kinase anchor protein 550
Protein rugose
dAKAP550
Protein rugose
dAKAP550
Location in the cell
PlasmaMembrane Reliability : 2.672
Sequence
CDS
ATGACGCAGAAGTGGCAGCGACGGGAGATCTCCAACTTCGAGTACCTTATGTTCCTGAATACCATTGCAGGTCGCACGTACAACGATCTGAACCAGTACCCCGTGTTCCCGTGGATCCTCACCAACTATGAGAGCGAAGAACTCGACCTTAGCCAGCCTTCAAACTTCAGGGATCTATCAAAGCCCATAGGTGCATTGAACCCCAGCCGACGTGCGTACTTCGAAGAGAGATATAACACGTGGGAGCACGAGTCCACGCCTCCCTTCCACTACGGCACACACTACTCCACTGCTGCTTTCGTGCTCAACTGGCTCGTTCGAATCGAGCCAATGACGACAATGTTCCTAGCGCTGCAGGGAGGTAAATTCGATCATCCGAACAGACTGTTTTCCTCCATTGCCATGTCCTGGAAGAACTGCCAACGCGACACATCCGATGTTAAGGAACTGATCCCAGAGCTGTTCTTCTTGCCTGAAATGCTGATAAACAGCTCCGGCTATAAGCTGGGCATCCCGGAGTCTCCGTCCGGCGACGTCATTCTTCCACCATGGGCTTCATCCCCTGAAGAGTTCATCCGCATCAACCGCATGGCCTTGGAGTCGGAGTTCGTCAGCTGCCAACTTCACCAGTGGATCGATCTCATCTTTGGATACAAACAGCGAGGGCCGGAAGCAGTACGGGCCACGAACGTGTTCTACTACCTCACGTACGAGGGCGGAGTCAGGCTCGACCAAATCACGGAGCCCGTGACCAGGACCGCCGTGGAGGACCAGATCCGCAGCTTCGGCCAGACCCCTGCGCAGCTACTGAGCGAGCCGCATCCGCCGCGCGCGTCTGCGCTACACCTGGCGCCGCTGATGTTCGCGGCGGCCCCCGACGACGTCTGTCTCCGCCTCAAGCTGCCCTCCAATGCGGCCGTCGTGCACGTCTCCGCCAACACCTACCCGCAGCTGGCGATGCCTGCTGCTGTCACGGTGAGCGCGGCGCGTGCCTTCGCGGCCCACCGCTGGGCGGCGGGGGGCTGCGCGCACTCCGCCCGCCACGCGCCCGACGCGCATCCGCCCCCCCACCCCCACCCGCTACTCATGGACCCAGTCTGGGCCGCTGGAGGAGCGCAGGCGCTGGCGCGGAGACAGCTCGGCGACAACTTCAGCCAGCGGGTCCGCGCGCGCTCAAACTGCTTCGTGACGACAGTCGACTCCAAGTTCCTCATCGCGGCCGGCTTCTGGGATAATAGCTTTAGGGTCTTCTCAACTGAAACTGCTAAAATAGTGCAAATAATATTCGGTCACTACGGCGTGGTGACGTGCGTTTGCCGGTCCGAGTGCAACATCACCTCGGACTGCTACATCGCGTCCGGCTCCGAGGACTGCACCGTGCTGCTCTGGCATTGGTCCGCCCGGCACGGCGGCATCGTCGGCGAGGGCGACAGTCCGGCGCCGCGCGTGACGCTCACGGGACACGACGCGCCCGTCAACGGCGTGCTCGTCTCTGCAGAACTCGGACTCGTCATCTCCTCCTCGCTCAACGGTCCGGTGCTGATCCACACGACGTTCGGCGAGGTGCTGCGCTCACTGAGCAGCGGGGAGGCGTCGGGTCCGCAGCAGCTGGCACTGTCCCGGGAGGGCGTGCTGGTGGTCGCGTACGCCAAGGGACACCTCGCCGCCTACACCCTCAACGGACGGAGGCTCAGGCATGAGACGCACAACGATAACTTCCAGGTAATCATTGCTACCGGAAGGCTACATGATGCAACGCGATGCCACTAA
Protein
MTQKWQRREISNFEYLMFLNTIAGRTYNDLNQYPVFPWILTNYESEELDLSQPSNFRDLSKPIGALNPSRRAYFEERYNTWEHESTPPFHYGTHYSTAAFVLNWLVRIEPMTTMFLALQGGKFDHPNRLFSSIAMSWKNCQRDTSDVKELIPELFFLPEMLINSSGYKLGIPESPSGDVILPPWASSPEEFIRINRMALESEFVSCQLHQWIDLIFGYKQRGPEAVRATNVFYYLTYEGGVRLDQITEPVTRTAVEDQIRSFGQTPAQLLSEPHPPRASALHLAPLMFAAAPDDVCLRLKLPSNAAVVHVSANTYPQLAMPAAVTVSAARAFAAHRWAAGGCAHSARHAPDAHPPPHPHPLLMDPVWAAGGAQALARRQLGDNFSQRVRARSNCFVTTVDSKFLIAAGFWDNSFRVFSTETAKIVQIIFGHYGVVTCVCRSECNITSDCYIASGSEDCTVLLWHWSARHGGIVGEGDSPAPRVTLTGHDAPVNGVLVSAELGLVISSSLNGPVLIHTTFGEVLRSLSSGEASGPQQLALSREGVLVVAYAKGHLAAYTLNGRRLRHETHNDNFQVIIATGRLHDATRCH
Summary
Description
Binds to type II regulatory subunits of protein kinase A and anchors/targets them to the membrane. May anchor the kinase to cytoskeletal and/or organelle-associated proteins. Required for correct retinal pattern formation and may function in cell fate determination through its interactions with the EGFR and Notch signaling pathways. Required for associative odor learning and short-term memory. Involved in development of the neuromuscular junction and the mushroom body.
Subunit
Interacts with RII subunit of PKA and components of the EGFR-mediated and Notch-mediated signaling pathways.
Similarity
Belongs to the WD repeat neurobeachin family.
Keywords
Alternative splicing
Complete proteome
Cytoplasm
Membrane
Reference proteome
Repeat
WD repeat
Feature
chain Neurobeachin
splice variant In isoform R.
splice variant In isoform R.
Uniprot
H9JIG7
A0A3S2NYH0
A0A2H1WTR7
A0A0N1PGJ6
A0A088AMU7
A0A158N9E3
+ More
A0A151JZB6 A0A195BGQ2 V9IKF6 A0A3L8DS32 A0A1B6FFQ9 A0A1B6FHD0 A0A1B6GFL2 A0A1B6GW90 E0VB80 K7J4I4 F4WJX1 A0A1B6M2V4 A0A1B6KZC9 A0A154PR26 A0A026WJE4 A0A1Y1KDW4 A0A146KQW0 A0A1Y1KFR1 A0A1Y1K804 A0A1Y1K827 A0A146M1B0 A0A067QFC6 E2AP05 A0A0L7R5X0 A0A0A9W0E0 A0A023F547 A0A224XGR8 A0A1W4XAT3 A0A1W4XBX6 A0A1W4X1F6 A0A1W4X1G1 A0A1W4X1B0 A0A1W4XBX0 A0A1W4XAS7 A0A1W4X1B5 A0A1W4XC55 A0A2J7PKZ6 D2A310 A0A2A3ERV9 A0A1B6DX44 A0A1B6DPZ3 A0A0T6BHE2 A0A1B0BVL9 A0A0K8TWW3 A0A1B0GJH6 A0A0R1E9Z0 A0A1B0G0Z8 X2JE60 A0A0R1E994 A0A1B0D3F4 A0A1I8M1Z6 A0A1I8M200 B7Z0X3 A0A0R1E9C2 A0A0R1EB30 B4MJ67 A0A1I8M202 Q0KHV9 A0A0R1E9K4 B4PZS6 A0A0R1E9M6 Q9W4E2 A0A1W4VI88 A0A1I8M203 A0A1I8M1Z8 A0A1I8M1Z5 A0A0Q5T5L6 B3NV21 Q9W4E2-6 X2JAH8 A0A0Q5T2R4 A0A0Q5TDD9 A0A0Q5THK5 A0A1W4V6H3 A0A1W4VI95 B7Z0W8 J9JVW6 A0A1I8Q9G7 A0A0Q5TID7 A0A0Q5T957 A0A0Q5T2S4 A0A1W4VIU7 A0A1W4V620 A0A1W4VJN1 A0A1W4VJM6 A0A1W4V635 A0A3B0K3L2 A0A3B0K3P9 A0A3B0K3M1 A0A1I8Q9G3 A0A1W4V6I2 A0A1W4VIV7 A0A1W4VIV2 A0A1W4V631
A0A151JZB6 A0A195BGQ2 V9IKF6 A0A3L8DS32 A0A1B6FFQ9 A0A1B6FHD0 A0A1B6GFL2 A0A1B6GW90 E0VB80 K7J4I4 F4WJX1 A0A1B6M2V4 A0A1B6KZC9 A0A154PR26 A0A026WJE4 A0A1Y1KDW4 A0A146KQW0 A0A1Y1KFR1 A0A1Y1K804 A0A1Y1K827 A0A146M1B0 A0A067QFC6 E2AP05 A0A0L7R5X0 A0A0A9W0E0 A0A023F547 A0A224XGR8 A0A1W4XAT3 A0A1W4XBX6 A0A1W4X1F6 A0A1W4X1G1 A0A1W4X1B0 A0A1W4XBX0 A0A1W4XAS7 A0A1W4X1B5 A0A1W4XC55 A0A2J7PKZ6 D2A310 A0A2A3ERV9 A0A1B6DX44 A0A1B6DPZ3 A0A0T6BHE2 A0A1B0BVL9 A0A0K8TWW3 A0A1B0GJH6 A0A0R1E9Z0 A0A1B0G0Z8 X2JE60 A0A0R1E994 A0A1B0D3F4 A0A1I8M1Z6 A0A1I8M200 B7Z0X3 A0A0R1E9C2 A0A0R1EB30 B4MJ67 A0A1I8M202 Q0KHV9 A0A0R1E9K4 B4PZS6 A0A0R1E9M6 Q9W4E2 A0A1W4VI88 A0A1I8M203 A0A1I8M1Z8 A0A1I8M1Z5 A0A0Q5T5L6 B3NV21 Q9W4E2-6 X2JAH8 A0A0Q5T2R4 A0A0Q5TDD9 A0A0Q5THK5 A0A1W4V6H3 A0A1W4VI95 B7Z0W8 J9JVW6 A0A1I8Q9G7 A0A0Q5TID7 A0A0Q5T957 A0A0Q5T2S4 A0A1W4VIU7 A0A1W4V620 A0A1W4VJN1 A0A1W4VJM6 A0A1W4V635 A0A3B0K3L2 A0A3B0K3P9 A0A3B0K3M1 A0A1I8Q9G3 A0A1W4V6I2 A0A1W4VIV7 A0A1W4VIV2 A0A1W4V631
Pubmed
19121390
26354079
21347285
30249741
20566863
20075255
+ More
21719571 24508170 28004739 26823975 24845553 20798317 25401762 25474469 18362917 19820115 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25315136 11102458 9334242 12537569 12072466 12124948 23100440 23790035
21719571 24508170 28004739 26823975 24845553 20798317 25401762 25474469 18362917 19820115 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25315136 11102458 9334242 12537569 12072466 12124948 23100440 23790035
EMBL
BABH01027226
BABH01027227
RSAL01000024
RVE52214.1
ODYU01010995
SOQ56460.1
+ More
KQ460817 KPJ12124.1 ADTU01009499 ADTU01009500 ADTU01009501 ADTU01009502 ADTU01009503 ADTU01009504 ADTU01009505 ADTU01009506 ADTU01009507 ADTU01009508 KQ981404 KYN41948.1 KQ976476 KYM83838.1 JR049953 AEY61137.1 QOIP01000005 RLU22983.1 GECZ01020723 JAS49046.1 GECZ01020160 JAS49609.1 GECZ01008527 JAS61242.1 GECZ01003156 JAS66613.1 DS235023 EEB10636.1 AAZX01002336 AAZX01003751 AAZX01006319 AAZX01008405 AAZX01009296 AAZX01009746 AAZX01011198 AAZX01012336 AAZX01015153 AAZX01016889 GL888187 EGI65493.1 GEBQ01009747 JAT30230.1 GEBQ01023170 JAT16807.1 KQ435066 KZC14375.1 KK107214 EZA55239.1 GEZM01089250 JAV57596.1 GDHC01021007 JAP97621.1 GEZM01089254 GEZM01089249 JAV57597.1 GEZM01089253 GEZM01089252 JAV57592.1 GEZM01089251 GEZM01089248 JAV57599.1 GDHC01005584 JAQ13045.1 KK853659 KDR02350.1 GL441439 EFN64837.1 KQ414648 KOC66214.1 GBHO01041707 JAG01897.1 GBBI01002524 JAC16188.1 GFTR01008776 JAW07650.1 NEVH01024529 PNF17001.1 KQ971338 EFA01972.2 KZ288190 PBC34533.1 GEDC01007044 JAS30254.1 GEDC01030294 GEDC01009543 JAS07004.1 JAS27755.1 LJIG01000188 KRT86752.1 JXJN01021367 JXJN01021368 GDHF01033362 JAI18952.1 AJWK01021725 AJWK01021726 AJWK01021727 AJWK01021728 AJWK01021729 AJWK01021730 AJWK01021731 AJWK01021732 AJWK01021733 AJWK01021734 CM000162 KRK05983.1 CCAG010000760 CCAG010000761 CCAG010000762 CCAG010000763 CCAG010000764 AE014298 AHN59349.1 KRK05984.1 AJVK01023522 AJVK01023523 AJVK01023524 ACL82890.2 KRK05980.1 KRK05982.1 CH963738 EDW72156.2 ABI30968.3 KRK05979.1 EDX01143.2 KRK05981.1 Y18278 AF003622 AY051596 CH954180 KQS29728.1 EDV45869.2 AHN59350.1 KQS29731.1 KQS29733.1 KQS29732.1 ACL82891.2 ABLF02025882 ABLF02025883 KQS29734.1 KQS29729.1 KQS29730.1 OUUW01000016 SPP88827.1 SPP88824.1 SPP88825.1
KQ460817 KPJ12124.1 ADTU01009499 ADTU01009500 ADTU01009501 ADTU01009502 ADTU01009503 ADTU01009504 ADTU01009505 ADTU01009506 ADTU01009507 ADTU01009508 KQ981404 KYN41948.1 KQ976476 KYM83838.1 JR049953 AEY61137.1 QOIP01000005 RLU22983.1 GECZ01020723 JAS49046.1 GECZ01020160 JAS49609.1 GECZ01008527 JAS61242.1 GECZ01003156 JAS66613.1 DS235023 EEB10636.1 AAZX01002336 AAZX01003751 AAZX01006319 AAZX01008405 AAZX01009296 AAZX01009746 AAZX01011198 AAZX01012336 AAZX01015153 AAZX01016889 GL888187 EGI65493.1 GEBQ01009747 JAT30230.1 GEBQ01023170 JAT16807.1 KQ435066 KZC14375.1 KK107214 EZA55239.1 GEZM01089250 JAV57596.1 GDHC01021007 JAP97621.1 GEZM01089254 GEZM01089249 JAV57597.1 GEZM01089253 GEZM01089252 JAV57592.1 GEZM01089251 GEZM01089248 JAV57599.1 GDHC01005584 JAQ13045.1 KK853659 KDR02350.1 GL441439 EFN64837.1 KQ414648 KOC66214.1 GBHO01041707 JAG01897.1 GBBI01002524 JAC16188.1 GFTR01008776 JAW07650.1 NEVH01024529 PNF17001.1 KQ971338 EFA01972.2 KZ288190 PBC34533.1 GEDC01007044 JAS30254.1 GEDC01030294 GEDC01009543 JAS07004.1 JAS27755.1 LJIG01000188 KRT86752.1 JXJN01021367 JXJN01021368 GDHF01033362 JAI18952.1 AJWK01021725 AJWK01021726 AJWK01021727 AJWK01021728 AJWK01021729 AJWK01021730 AJWK01021731 AJWK01021732 AJWK01021733 AJWK01021734 CM000162 KRK05983.1 CCAG010000760 CCAG010000761 CCAG010000762 CCAG010000763 CCAG010000764 AE014298 AHN59349.1 KRK05984.1 AJVK01023522 AJVK01023523 AJVK01023524 ACL82890.2 KRK05980.1 KRK05982.1 CH963738 EDW72156.2 ABI30968.3 KRK05979.1 EDX01143.2 KRK05981.1 Y18278 AF003622 AY051596 CH954180 KQS29728.1 EDV45869.2 AHN59350.1 KQS29731.1 KQS29733.1 KQS29732.1 ACL82891.2 ABLF02025882 ABLF02025883 KQS29734.1 KQS29729.1 KQS29730.1 OUUW01000016 SPP88827.1 SPP88824.1 SPP88825.1
Proteomes
UP000005204
UP000283053
UP000053240
UP000005203
UP000005205
UP000078541
+ More
UP000078540 UP000279307 UP000009046 UP000002358 UP000007755 UP000076502 UP000053097 UP000027135 UP000000311 UP000053825 UP000192223 UP000235965 UP000007266 UP000242457 UP000092460 UP000092461 UP000002282 UP000092444 UP000000803 UP000092462 UP000095301 UP000007798 UP000192221 UP000008711 UP000007819 UP000095300 UP000268350
UP000078540 UP000279307 UP000009046 UP000002358 UP000007755 UP000076502 UP000053097 UP000027135 UP000000311 UP000053825 UP000192223 UP000235965 UP000007266 UP000242457 UP000092460 UP000092461 UP000002282 UP000092444 UP000000803 UP000092462 UP000095301 UP000007798 UP000192221 UP000008711 UP000007819 UP000095300 UP000268350
Interpro
IPR015943
WD40/YVTN_repeat-like_dom_sf
+ More
IPR036372 BEACH_dom_sf
IPR001680 WD40_repeat
IPR000409 BEACH_dom
IPR036322 WD40_repeat_dom_sf
IPR017986 WD40_repeat_dom
IPR023362 PH-BEACH_dom
IPR010508 DUF1088
IPR016024 ARM-type_fold
IPR031570 DUF4704
IPR013320 ConA-like_dom_sf
IPR000253 FHA_dom
IPR001611 Leu-rich_rpt
IPR036372 BEACH_dom_sf
IPR001680 WD40_repeat
IPR000409 BEACH_dom
IPR036322 WD40_repeat_dom_sf
IPR017986 WD40_repeat_dom
IPR023362 PH-BEACH_dom
IPR010508 DUF1088
IPR016024 ARM-type_fold
IPR031570 DUF4704
IPR013320 ConA-like_dom_sf
IPR000253 FHA_dom
IPR001611 Leu-rich_rpt
Gene 3D
ProteinModelPortal
H9JIG7
A0A3S2NYH0
A0A2H1WTR7
A0A0N1PGJ6
A0A088AMU7
A0A158N9E3
+ More
A0A151JZB6 A0A195BGQ2 V9IKF6 A0A3L8DS32 A0A1B6FFQ9 A0A1B6FHD0 A0A1B6GFL2 A0A1B6GW90 E0VB80 K7J4I4 F4WJX1 A0A1B6M2V4 A0A1B6KZC9 A0A154PR26 A0A026WJE4 A0A1Y1KDW4 A0A146KQW0 A0A1Y1KFR1 A0A1Y1K804 A0A1Y1K827 A0A146M1B0 A0A067QFC6 E2AP05 A0A0L7R5X0 A0A0A9W0E0 A0A023F547 A0A224XGR8 A0A1W4XAT3 A0A1W4XBX6 A0A1W4X1F6 A0A1W4X1G1 A0A1W4X1B0 A0A1W4XBX0 A0A1W4XAS7 A0A1W4X1B5 A0A1W4XC55 A0A2J7PKZ6 D2A310 A0A2A3ERV9 A0A1B6DX44 A0A1B6DPZ3 A0A0T6BHE2 A0A1B0BVL9 A0A0K8TWW3 A0A1B0GJH6 A0A0R1E9Z0 A0A1B0G0Z8 X2JE60 A0A0R1E994 A0A1B0D3F4 A0A1I8M1Z6 A0A1I8M200 B7Z0X3 A0A0R1E9C2 A0A0R1EB30 B4MJ67 A0A1I8M202 Q0KHV9 A0A0R1E9K4 B4PZS6 A0A0R1E9M6 Q9W4E2 A0A1W4VI88 A0A1I8M203 A0A1I8M1Z8 A0A1I8M1Z5 A0A0Q5T5L6 B3NV21 Q9W4E2-6 X2JAH8 A0A0Q5T2R4 A0A0Q5TDD9 A0A0Q5THK5 A0A1W4V6H3 A0A1W4VI95 B7Z0W8 J9JVW6 A0A1I8Q9G7 A0A0Q5TID7 A0A0Q5T957 A0A0Q5T2S4 A0A1W4VIU7 A0A1W4V620 A0A1W4VJN1 A0A1W4VJM6 A0A1W4V635 A0A3B0K3L2 A0A3B0K3P9 A0A3B0K3M1 A0A1I8Q9G3 A0A1W4V6I2 A0A1W4VIV7 A0A1W4VIV2 A0A1W4V631
A0A151JZB6 A0A195BGQ2 V9IKF6 A0A3L8DS32 A0A1B6FFQ9 A0A1B6FHD0 A0A1B6GFL2 A0A1B6GW90 E0VB80 K7J4I4 F4WJX1 A0A1B6M2V4 A0A1B6KZC9 A0A154PR26 A0A026WJE4 A0A1Y1KDW4 A0A146KQW0 A0A1Y1KFR1 A0A1Y1K804 A0A1Y1K827 A0A146M1B0 A0A067QFC6 E2AP05 A0A0L7R5X0 A0A0A9W0E0 A0A023F547 A0A224XGR8 A0A1W4XAT3 A0A1W4XBX6 A0A1W4X1F6 A0A1W4X1G1 A0A1W4X1B0 A0A1W4XBX0 A0A1W4XAS7 A0A1W4X1B5 A0A1W4XC55 A0A2J7PKZ6 D2A310 A0A2A3ERV9 A0A1B6DX44 A0A1B6DPZ3 A0A0T6BHE2 A0A1B0BVL9 A0A0K8TWW3 A0A1B0GJH6 A0A0R1E9Z0 A0A1B0G0Z8 X2JE60 A0A0R1E994 A0A1B0D3F4 A0A1I8M1Z6 A0A1I8M200 B7Z0X3 A0A0R1E9C2 A0A0R1EB30 B4MJ67 A0A1I8M202 Q0KHV9 A0A0R1E9K4 B4PZS6 A0A0R1E9M6 Q9W4E2 A0A1W4VI88 A0A1I8M203 A0A1I8M1Z8 A0A1I8M1Z5 A0A0Q5T5L6 B3NV21 Q9W4E2-6 X2JAH8 A0A0Q5T2R4 A0A0Q5TDD9 A0A0Q5THK5 A0A1W4V6H3 A0A1W4VI95 B7Z0W8 J9JVW6 A0A1I8Q9G7 A0A0Q5TID7 A0A0Q5T957 A0A0Q5T2S4 A0A1W4VIU7 A0A1W4V620 A0A1W4VJN1 A0A1W4VJM6 A0A1W4V635 A0A3B0K3L2 A0A3B0K3P9 A0A3B0K3M1 A0A1I8Q9G3 A0A1W4V6I2 A0A1W4VIV7 A0A1W4VIV2 A0A1W4V631
PDB
1MI1
E-value=2.27491e-129,
Score=1185
Ontologies
GO
Topology
Subcellular location
Cytoplasm
Membrane
Perikaryon
Membrane
Perikaryon
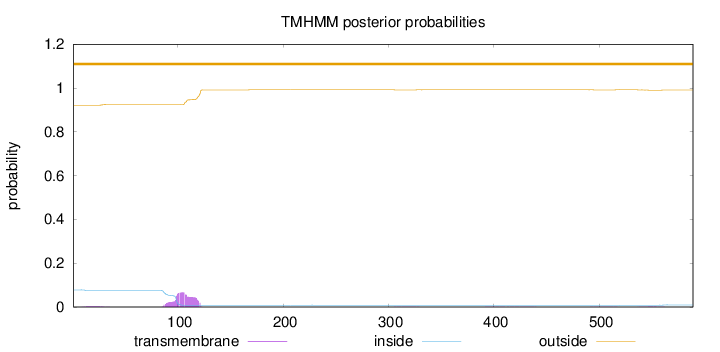
Length:
589
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.51525
Exp number, first 60 AAs:
0.05316
Total prob of N-in:
0.07819
outside
1 - 589
Population Genetic Test Statistics
Pi
232.838996
Theta
162.344986
Tajima's D
1.272656
CLR
0.064906
CSRT
0.730213489325534
Interpretation
Uncertain
