Gene
KWMTBOMO08543 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009319
Annotation
ribosomal_protein_S3_[Bombyx_mori]
Full name
40S ribosomal protein S3
+ More
40S ribosomal protein S3-A
40S ribosomal protein S3-A
Alternative Name
S1A
Location in the cell
Cytoplasmic Reliability : 1.97 Mitochondrial Reliability : 1.666
Sequence
CDS
ATGGCCGTGAACAATATTTCAAAGAAGCGAAAATTTGTTGGAGATGGAGTTTTCAAGGCGGAACTCAATGAGTTCCTCACTAGGGAGCTGGCCGAGGACGGCTACTCCGGCGTGGAAGTCCGGGTCACTCCCATCCGCTCGGAGATCATTATTATGGCCACCAGGACACAAAGTGTGCTCGGAGAGAAAGGACGCAGAATCCGTGAGCTGACTTCCGTAGTACAGAAGCGATTCAACATTCCAGAGCAATCTGTAGAATTGTATGCTGAAAAGGTGGCTACTCGTGGTCTTTGCGCTATCGCCCAGGCCGAATCTCTAAGATACAAGCTTATCGGAGGTCTCGCTGTACGTCGTGCTTGCTATGGTGTTCTCCGTTTCATCATGGAATCTGGCGCCCGTGGTTGTGAAGTTGTTGTATCTGGCAAGCTGCGTGGTCAACGTGCCAAATCAATGAAGTTTGTAGATGGACTCATGATCCACTCTGGAGACCCTTGCAATGACTACGTCAACACTGCTACCAGACATGTGCTTCTCAGGCAAGGAGTACTAGGAATCAAGGTCAAAATCATGTTGCCGTGGGACCAGCAAGGCAAGAACGGCCCGAAGAAGCCACAACCAGACCACATCCTGGTGACAGAGCCCAAGGACGAGCCCGTGCCCCTCGAGCCGACCAGCGAAGTGCGCTCCCTCGCGCCCGCGCCTCTGCCGCAGCCGGTCGCCGCTGTCGCTTAG
Protein
MAVNNISKKRKFVGDGVFKAELNEFLTRELAEDGYSGVEVRVTPIRSEIIIMATRTQSVLGEKGRRIRELTSVVQKRFNIPEQSVELYAEKVATRGLCAIAQAESLRYKLIGGLAVRRACYGVLRFIMESGARGCEVVVSGKLRGQRAKSMKFVDGLMIHSGDPCNDYVNTATRHVLLRQGVLGIKVKIMLPWDQQGKNGPKKPQPDHILVTEPKDEPVPLEPTSEVRSLAPAPLPQPVAAVA
Summary
Description
Involved in translation as a component of the 40S small ribosomal subunit. Has endonuclease activity and plays a role in repair of damaged DNA. Also involved in other processes including regulation of transcription, translation of its cognate mRNA, spindle formation and chromosome movement during mitosis, and apoptosis.
Catalytic Activity
The C-O-P bond 3' to the apurinic or apyrimidinic site in DNA is broken by a beta-elimination reaction, leaving a 3'-terminal unsaturated sugar and a product with a terminal 5'-phosphate.
Similarity
Belongs to the universal ribosomal protein uS3 family.
Keywords
Ribonucleoprotein
Ribosomal protein
RNA-binding
Apoptosis
Cell cycle
Cell division
Cytoplasm
Cytoskeleton
DNA damage
DNA repair
DNA-binding
Lyase
Membrane
Mitochondrion
Mitochondrion inner membrane
Mitosis
Nucleus
Transcription
Transcription regulation
Translation regulation
Feature
chain 40S ribosomal protein S3
Uniprot
Q5UAP2
A0A2H1WTQ8
E7DYZ6
A0A3G1T1G0
A0A089RLZ6
A0A0N1IP25
+ More
I4DIK4 I4DLY2 P48153 A0A0A1E4H3 Q95V36 A0A212FND8 G0ZEB3 Q000A7 A0A0L7LNM2 A0A3S2LEV6 Q6EV28 Q4GXR1 A0A0N9MP84 A0A1W4XWB0 A0A146KR45 A0A1B1NJ06 A0A1Y1LA29 A0A1D1XCH0 A0A067R1U8 A0A2P8XV80 Q6EV05 A0A2J7R3C1 A0A0U2TI96 K7J8W6 R4V2V4 A2IA98 A0A1B0GIF5 A0A0C9QZ78 A0A1B6M639 A0A0X8NRC9 A0A0U2V6K3 A0A154NXU5 Q56FH9 A0A026W1G3 A0A077CUU1 D1M7X5 A0A1L8HJY0 P02350 Q7ZYT3 V5H4W7 A0A3Q2DY75 A0A1B1MRN5 A0A0U2T8D0 A0A0U2UT23 G3PJC8 U5EUR5 M1K4Y5 B5DGY5 W5MH79 C0H7T0 B5XAC5 A0A3B3XT32 A0A3B5Q286 A0A3B5LZC5 A0A087Y773 A0A3B3VA71 A0A3P9PWI0 A0A1L8DSD4 A0A0S7MHH5 A0A060VTT3 A0A2V2PN55 A0A3B3QSN6 A0A0K2UH90 A0A1B6GTJ0 A0A0U2UH62 A0A1S3LP02 A0A3P8ZA90 E2BHI4 A0A1A8FTZ9 A0A1A7YWJ2 A0A0E9WR13 A0A0B8RP95 A0A060WYE0 A0A1A8GP69 K9J1W0 F6RG35 Q6QZI0 A0A088AV85 A0A1W7R9N7 A0A1A8P4X7 A0A1A8Q4S8 A0A1A8I5S5 A0A1A8E8E0 A0A1A8UXU3 A0A348G678 A0A1S3F049 A0A3L7HXG6 I3N0Y3 A0A1U7Q9Q4 A0A250YDI8 A0A286XTB7 A0A091D1I9 Q5YLW3 L5JLX8
I4DIK4 I4DLY2 P48153 A0A0A1E4H3 Q95V36 A0A212FND8 G0ZEB3 Q000A7 A0A0L7LNM2 A0A3S2LEV6 Q6EV28 Q4GXR1 A0A0N9MP84 A0A1W4XWB0 A0A146KR45 A0A1B1NJ06 A0A1Y1LA29 A0A1D1XCH0 A0A067R1U8 A0A2P8XV80 Q6EV05 A0A2J7R3C1 A0A0U2TI96 K7J8W6 R4V2V4 A2IA98 A0A1B0GIF5 A0A0C9QZ78 A0A1B6M639 A0A0X8NRC9 A0A0U2V6K3 A0A154NXU5 Q56FH9 A0A026W1G3 A0A077CUU1 D1M7X5 A0A1L8HJY0 P02350 Q7ZYT3 V5H4W7 A0A3Q2DY75 A0A1B1MRN5 A0A0U2T8D0 A0A0U2UT23 G3PJC8 U5EUR5 M1K4Y5 B5DGY5 W5MH79 C0H7T0 B5XAC5 A0A3B3XT32 A0A3B5Q286 A0A3B5LZC5 A0A087Y773 A0A3B3VA71 A0A3P9PWI0 A0A1L8DSD4 A0A0S7MHH5 A0A060VTT3 A0A2V2PN55 A0A3B3QSN6 A0A0K2UH90 A0A1B6GTJ0 A0A0U2UH62 A0A1S3LP02 A0A3P8ZA90 E2BHI4 A0A1A8FTZ9 A0A1A7YWJ2 A0A0E9WR13 A0A0B8RP95 A0A060WYE0 A0A1A8GP69 K9J1W0 F6RG35 Q6QZI0 A0A088AV85 A0A1W7R9N7 A0A1A8P4X7 A0A1A8Q4S8 A0A1A8I5S5 A0A1A8E8E0 A0A1A8UXU3 A0A348G678 A0A1S3F049 A0A3L7HXG6 I3N0Y3 A0A1U7Q9Q4 A0A250YDI8 A0A286XTB7 A0A091D1I9 Q5YLW3 L5JLX8
EC Number
4.2.99.18
Pubmed
19121390
27163122
26354079
22651552
8630533
14668217
+ More
22118469 16885282 26227816 26823975 28004739 24845553 29403074 26621068 20075255 17437641 24508170 30249741 27762356 2030971 7049839 19878547 20433749 23542700 24755649 29240929 25069045 20798317 25613341 25476704 17495919 15792627 29704459 28087693 21993624 10349636 11042159 11076861 11217851 12466851 12040188 16141073 16356660 17242355 19144319 21183079 23806337 23258410
22118469 16885282 26227816 26823975 28004739 24845553 29403074 26621068 20075255 17437641 24508170 30249741 27762356 2030971 7049839 19878547 20433749 23542700 24755649 29240929 25069045 20798317 25613341 25476704 17495919 15792627 29704459 28087693 21993624 10349636 11042159 11076861 11217851 12466851 12040188 16141073 16356660 17242355 19144319 21183079 23806337 23258410
EMBL
BABH01027225
AY769316
AAV34858.1
ODYU01010995
SOQ56459.1
HQ424699
+ More
ADT80643.1 MG992382 AXY94820.1 KM064630 KT962961 AIR07416.1 AND95944.1 KQ460817 KPJ12125.1 AK401122 KQ459337 BAM17744.1 KPJ01370.1 AK402300 BAM18922.1 U12708 KJ405954 AIY22794.1 AF429976 AAL26578.1 AGBW02006209 OWR55233.1 JF264991 AEL28822.1 DQ988989 ABJ55992.1 JTDY01000523 KOB76806.1 RSAL01000024 RVE52215.1 AJ783759 CAH04122.1 AM048962 CAJ17197.1 KR080494 ALG76022.1 GDHC01020902 JAP97726.1 KT274801 ANS83636.1 GEZM01063261 JAV69160.1 GDJX01027940 JAT39996.1 KK852773 KDR16813.1 PYGN01001295 PSN35918.1 AJ783866 CAH04314.1 NEVH01007822 PNF35323.1 KT755242 ALS05076.1 AAZX01000813 AAZX01013133 KC632284 AGM32098.1 EF179412 ABM55418.1 AJWK01014059 GBYB01006002 JAG75769.1 GEBQ01008623 JAT31354.1 KC507300 AHB12423.1 KT754746 KT754747 ALS04581.1 KQ434778 KZC04433.1 AY961521 AAX62423.1 KK107488 QOIP01000012 EZA49902.1 RLU15845.1 KJ868729 AIL23530.1 GU120399 ACY71249.1 CM004468 OCT96376.1 X57322 BC042230 V01441 BC041299 BC169756 BC169758 CM004469 AAH41299.1 AAI69756.1 OCT93347.1 GALX01000613 JAB67853.1 KU521367 ANS71247.1 KT755094 ALS04928.1 KT754933 ALS04767.1 GANO01002203 JAB57668.1 KC109783 AGF29412.1 BT043894 BT047476 BT058725 ACH71009.1 ACI67277.1 ACN10438.1 AHAT01003067 BT058386 BT060087 ACN10099.1 ACN12447.1 BT047994 ACI67795.1 AYCK01004889 GFDF01004773 JAV09311.1 GBYX01024237 JAP01208.1 FR904302 CDQ58373.1 QGNU01000090 PWS23245.1 HACA01020212 CDW37573.1 GECZ01004106 JAS65663.1 KT755329 ALS05163.1 GL448301 EFN84837.1 HAEB01016348 SBQ62875.1 HADX01012745 SBP34977.1 GBXM01015780 JAH92797.1 GBSH01002387 JAG66639.1 FR904677 CDQ69610.1 HAEC01004117 SBQ72194.1 GABZ01002484 JAA51041.1 AY521669 AAT01919.1 GFAH01000533 JAV47856.1 HAEI01001427 SBR76129.1 HAEF01004352 HAEG01010947 SBR88458.1 HAED01005343 HAEE01014299 SBQ91373.1 HADZ01001902 HAEA01012889 SBQ41369.1 HADY01019651 HAEJ01012762 SBS53219.1 FX985620 BBF97951.1 RAZU01000137 RLQ70634.1 AGTP01028900 GFFW01003189 JAV41599.1 AAKN02051174 KN123669 KFO24040.1 AY043296 AK003181 AK010579 AK012234 AK051243 AK146796 AK148030 AK159508 AK160526 AK164077 AK165174 AK166750 AK167210 AK167266 AK168845 CH466531 AAK95377.1 BAB22624.1 BAC34570.1 BAE27438.1 BAE35844.1 BAE37615.1 BAE40668.1 EDL16392.1 KB031158 ELK00374.1
ADT80643.1 MG992382 AXY94820.1 KM064630 KT962961 AIR07416.1 AND95944.1 KQ460817 KPJ12125.1 AK401122 KQ459337 BAM17744.1 KPJ01370.1 AK402300 BAM18922.1 U12708 KJ405954 AIY22794.1 AF429976 AAL26578.1 AGBW02006209 OWR55233.1 JF264991 AEL28822.1 DQ988989 ABJ55992.1 JTDY01000523 KOB76806.1 RSAL01000024 RVE52215.1 AJ783759 CAH04122.1 AM048962 CAJ17197.1 KR080494 ALG76022.1 GDHC01020902 JAP97726.1 KT274801 ANS83636.1 GEZM01063261 JAV69160.1 GDJX01027940 JAT39996.1 KK852773 KDR16813.1 PYGN01001295 PSN35918.1 AJ783866 CAH04314.1 NEVH01007822 PNF35323.1 KT755242 ALS05076.1 AAZX01000813 AAZX01013133 KC632284 AGM32098.1 EF179412 ABM55418.1 AJWK01014059 GBYB01006002 JAG75769.1 GEBQ01008623 JAT31354.1 KC507300 AHB12423.1 KT754746 KT754747 ALS04581.1 KQ434778 KZC04433.1 AY961521 AAX62423.1 KK107488 QOIP01000012 EZA49902.1 RLU15845.1 KJ868729 AIL23530.1 GU120399 ACY71249.1 CM004468 OCT96376.1 X57322 BC042230 V01441 BC041299 BC169756 BC169758 CM004469 AAH41299.1 AAI69756.1 OCT93347.1 GALX01000613 JAB67853.1 KU521367 ANS71247.1 KT755094 ALS04928.1 KT754933 ALS04767.1 GANO01002203 JAB57668.1 KC109783 AGF29412.1 BT043894 BT047476 BT058725 ACH71009.1 ACI67277.1 ACN10438.1 AHAT01003067 BT058386 BT060087 ACN10099.1 ACN12447.1 BT047994 ACI67795.1 AYCK01004889 GFDF01004773 JAV09311.1 GBYX01024237 JAP01208.1 FR904302 CDQ58373.1 QGNU01000090 PWS23245.1 HACA01020212 CDW37573.1 GECZ01004106 JAS65663.1 KT755329 ALS05163.1 GL448301 EFN84837.1 HAEB01016348 SBQ62875.1 HADX01012745 SBP34977.1 GBXM01015780 JAH92797.1 GBSH01002387 JAG66639.1 FR904677 CDQ69610.1 HAEC01004117 SBQ72194.1 GABZ01002484 JAA51041.1 AY521669 AAT01919.1 GFAH01000533 JAV47856.1 HAEI01001427 SBR76129.1 HAEF01004352 HAEG01010947 SBR88458.1 HAED01005343 HAEE01014299 SBQ91373.1 HADZ01001902 HAEA01012889 SBQ41369.1 HADY01019651 HAEJ01012762 SBS53219.1 FX985620 BBF97951.1 RAZU01000137 RLQ70634.1 AGTP01028900 GFFW01003189 JAV41599.1 AAKN02051174 KN123669 KFO24040.1 AY043296 AK003181 AK010579 AK012234 AK051243 AK146796 AK148030 AK159508 AK160526 AK164077 AK165174 AK166750 AK167210 AK167266 AK168845 CH466531 AAK95377.1 BAB22624.1 BAC34570.1 BAE27438.1 BAE35844.1 BAE37615.1 BAE40668.1 EDL16392.1 KB031158 ELK00374.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000037510
UP000283053
+ More
UP000192223 UP000027135 UP000245037 UP000235965 UP000002358 UP000092461 UP000076502 UP000053097 UP000279307 UP000186698 UP000265020 UP000007635 UP000087266 UP000018468 UP000261480 UP000002852 UP000261380 UP000028760 UP000261500 UP000242638 UP000193380 UP000245538 UP000261540 UP000265140 UP000008237 UP000002280 UP000005203 UP000081671 UP000273346 UP000005215 UP000189706 UP000005447 UP000028990 UP000010552
UP000192223 UP000027135 UP000245037 UP000235965 UP000002358 UP000092461 UP000076502 UP000053097 UP000279307 UP000186698 UP000265020 UP000007635 UP000087266 UP000018468 UP000261480 UP000002852 UP000261380 UP000028760 UP000261500 UP000242638 UP000193380 UP000245538 UP000261540 UP000265140 UP000008237 UP000002280 UP000005203 UP000081671 UP000273346 UP000005215 UP000189706 UP000005447 UP000028990 UP000010552
PRIDE
Interpro
Gene 3D
ProteinModelPortal
Q5UAP2
A0A2H1WTQ8
E7DYZ6
A0A3G1T1G0
A0A089RLZ6
A0A0N1IP25
+ More
I4DIK4 I4DLY2 P48153 A0A0A1E4H3 Q95V36 A0A212FND8 G0ZEB3 Q000A7 A0A0L7LNM2 A0A3S2LEV6 Q6EV28 Q4GXR1 A0A0N9MP84 A0A1W4XWB0 A0A146KR45 A0A1B1NJ06 A0A1Y1LA29 A0A1D1XCH0 A0A067R1U8 A0A2P8XV80 Q6EV05 A0A2J7R3C1 A0A0U2TI96 K7J8W6 R4V2V4 A2IA98 A0A1B0GIF5 A0A0C9QZ78 A0A1B6M639 A0A0X8NRC9 A0A0U2V6K3 A0A154NXU5 Q56FH9 A0A026W1G3 A0A077CUU1 D1M7X5 A0A1L8HJY0 P02350 Q7ZYT3 V5H4W7 A0A3Q2DY75 A0A1B1MRN5 A0A0U2T8D0 A0A0U2UT23 G3PJC8 U5EUR5 M1K4Y5 B5DGY5 W5MH79 C0H7T0 B5XAC5 A0A3B3XT32 A0A3B5Q286 A0A3B5LZC5 A0A087Y773 A0A3B3VA71 A0A3P9PWI0 A0A1L8DSD4 A0A0S7MHH5 A0A060VTT3 A0A2V2PN55 A0A3B3QSN6 A0A0K2UH90 A0A1B6GTJ0 A0A0U2UH62 A0A1S3LP02 A0A3P8ZA90 E2BHI4 A0A1A8FTZ9 A0A1A7YWJ2 A0A0E9WR13 A0A0B8RP95 A0A060WYE0 A0A1A8GP69 K9J1W0 F6RG35 Q6QZI0 A0A088AV85 A0A1W7R9N7 A0A1A8P4X7 A0A1A8Q4S8 A0A1A8I5S5 A0A1A8E8E0 A0A1A8UXU3 A0A348G678 A0A1S3F049 A0A3L7HXG6 I3N0Y3 A0A1U7Q9Q4 A0A250YDI8 A0A286XTB7 A0A091D1I9 Q5YLW3 L5JLX8
I4DIK4 I4DLY2 P48153 A0A0A1E4H3 Q95V36 A0A212FND8 G0ZEB3 Q000A7 A0A0L7LNM2 A0A3S2LEV6 Q6EV28 Q4GXR1 A0A0N9MP84 A0A1W4XWB0 A0A146KR45 A0A1B1NJ06 A0A1Y1LA29 A0A1D1XCH0 A0A067R1U8 A0A2P8XV80 Q6EV05 A0A2J7R3C1 A0A0U2TI96 K7J8W6 R4V2V4 A2IA98 A0A1B0GIF5 A0A0C9QZ78 A0A1B6M639 A0A0X8NRC9 A0A0U2V6K3 A0A154NXU5 Q56FH9 A0A026W1G3 A0A077CUU1 D1M7X5 A0A1L8HJY0 P02350 Q7ZYT3 V5H4W7 A0A3Q2DY75 A0A1B1MRN5 A0A0U2T8D0 A0A0U2UT23 G3PJC8 U5EUR5 M1K4Y5 B5DGY5 W5MH79 C0H7T0 B5XAC5 A0A3B3XT32 A0A3B5Q286 A0A3B5LZC5 A0A087Y773 A0A3B3VA71 A0A3P9PWI0 A0A1L8DSD4 A0A0S7MHH5 A0A060VTT3 A0A2V2PN55 A0A3B3QSN6 A0A0K2UH90 A0A1B6GTJ0 A0A0U2UH62 A0A1S3LP02 A0A3P8ZA90 E2BHI4 A0A1A8FTZ9 A0A1A7YWJ2 A0A0E9WR13 A0A0B8RP95 A0A060WYE0 A0A1A8GP69 K9J1W0 F6RG35 Q6QZI0 A0A088AV85 A0A1W7R9N7 A0A1A8P4X7 A0A1A8Q4S8 A0A1A8I5S5 A0A1A8E8E0 A0A1A8UXU3 A0A348G678 A0A1S3F049 A0A3L7HXG6 I3N0Y3 A0A1U7Q9Q4 A0A250YDI8 A0A286XTB7 A0A091D1I9 Q5YLW3 L5JLX8
PDB
6MTE
E-value=5.85526e-105,
Score=970
Ontologies
GO
GO:0015935
GO:0003723
GO:0006412
GO:0003735
GO:0005840
GO:0004364
GO:0051301
GO:0007049
GO:0006281
GO:0006915
GO:0006417
GO:0005743
GO:0005819
GO:0005730
GO:0003677
GO:0140078
GO:0030218
GO:0043009
GO:0045739
GO:0022627
GO:0003684
GO:0008534
GO:0003906
GO:2001235
GO:0005634
GO:0032357
GO:0051225
GO:0050862
GO:0005783
GO:0070301
GO:0044877
GO:0032587
GO:0005844
GO:1905053
GO:0032358
GO:0044390
GO:0000977
GO:0014069
GO:0061481
GO:0071356
GO:0072686
GO:0042769
GO:0008017
GO:0051092
GO:0032079
GO:0071159
GO:0070181
GO:0031397
GO:1902231
GO:0097100
GO:0031116
GO:0008134
GO:0003729
GO:0051879
GO:0010628
GO:0032743
GO:0051018
GO:0042104
GO:0017148
GO:1901224
GO:0007059
GO:0043507
GO:2001272
GO:0019901
GO:0030544
GO:0045738
GO:0005759
GO:1902546
GO:0004520
GO:0004812
GO:0006418
GO:0003676
GO:0016876
GO:0043039
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleolus
Mitochondrion inner membrane
Cytoskeleton
Spindle
Nucleus
Nucleolus
Mitochondrion inner membrane
Cytoskeleton
Spindle
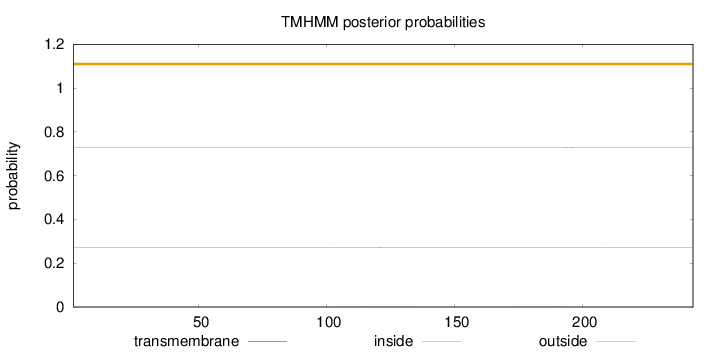
Length:
243
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01711
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.27291
outside
1 - 243
Population Genetic Test Statistics
Pi
427.381946
Theta
224.626624
Tajima's D
2.867981
CLR
0.136118
CSRT
0.970001499925004
Interpretation
Uncertain
