Gene
KWMTBOMO08542 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009375
Annotation
PREDICTED:_Krueppel_homolog_2_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.913
Sequence
CDS
ATGGCCGATCAAAATCAGCAAGCAGGCGACACTGGTCCACCAAAAGGCATCCCAGCCTTGAAAGCTCACATCATTGCCAATAAAATAGACGTAGCTCTTTGGGGGGTCCGCGTAATCACCGTGCTGTGTACCATTGGATACGTGTTCCCATTGTTTAACAATCCAGTGTCGGCATTCTACAAAGCGTTACTCGCGAACGCTGCGACTTCAGCGCTGCGGCTACATCAAAGAATTCCAGCACGAGAGATCTCCCTCTCGCGGGATTTCATGGCGAGGTTCTTCCTGGAGGACAGCGCTCACTATTTATTCTATTCACTCATTTTCATGAACGTCGTGCCCAATTTGTTAATATTGGTGCCAATCTTTTTATTTGCGCTGCTACACGCCGCTTCGTATTCCTTGACTATCCTTGATACTCTTGGTCAAAATTCGTTGTGGGTGGCTCGTCTGCTCATCTCCCTGGTCGAGTTCCAGTCGCGGAACATCCTCCGGGCCGCGGCGCTCGCCGAGATCGTGCTGTTCCCGGTCGTACTCATCATGGCATTGTTCGGTCACTGCGGCTTGATGAGTCCGTTCGTATACTACTACTTCGTGACGTGGAGGTACGCGTCGCGCCGGAACCCGTACACCCGCAACACGTTCCGCGAGCTCCGCGTGGCCGCCGACGGACTCACGCAGCGCCCCTGGGTGCCCGCCGCCCTCGCCTCCGCGCTGCGCTCCGCCATCGACATCGTGTGCCGCATGGCGCCGCCCGTCGAGCAACAACAACAGTAA
Protein
MADQNQQAGDTGPPKGIPALKAHIIANKIDVALWGVRVITVLCTIGYVFPLFNNPVSAFYKALLANAATSALRLHQRIPAREISLSRDFMARFFLEDSAHYLFYSLIFMNVVPNLLILVPIFLFALLHAASYSLTILDTLGQNSLWVARLLISLVEFQSRNILRAAALAEIVLFPVVLIMALFGHCGLMSPFVYYYFVTWRYASRRNPYTRNTFRELRVAADGLTQRPWVPAALASALRSAIDIVCRMAPPVEQQQQ
Summary
Uniprot
A0A2H1WTS4
A0A437BP80
S4PAU2
A0A194QD43
A0A212FNB1
A0A1E1WNI1
+ More
A0A1B0CSN0 A0A067R3Y8 A0A1L8D952 U5EI74 A0A1L8D9F2 A0A023ELE4 A0A0P6J696 A0A1B6K020 A0A2J7R3A5 A0A336KH59 B0WUC5 A0A1B6MTT8 A0A1Q3FGJ7 A0A1Q3FGJ9 J9JWN4 A0A2P8XV88 T1D4J6 T1E296 A0A2H8TUW5 A0A2S2QJC4 A0A1J1IQF6 A0A069DRJ7 A0A224XID3 T1DLV6 A0A2M3ZG19 W5JM07 A0A1I8JSS7 A0A2M4A3M5 A0A026W2T4 A0A1Q3FGQ1 A0A0V0G5T1 A0A084WD32 A0A034W1F0 A0A0C9RCI8 A0A1Y1KAB1 A0A023F749 N6TWS6 A0A2M4BX00 A0A182JMP6 A0A1W4XFU2 F4WME4 V5GWR6 E0VDR3 A0A158P1X0 A0A1I8JW70 A0A1S4H3B3 A0A1I8JTQ2 A0A182V837 A0A0A1WIU5 A0A0L0CJT7 A0A182KJS3 A0A195BSZ2 Q7PJT0 E2AZJ5 A0A182K0A5 A0A1I8Q3I0 A0A0M8ZW82 E2BHI3 A0A1L8EDW8 A0A195D233 A0A182Q308 R4WDR1 A0A3R7MJV6 A0A1B0A856 A0A182M7T5 A0A0P4WTN1 A0A1L8E940 A0A0K8WK73 A0A0K8UIB5 A0A0K8TQM7 A0A1B6DU58 A0A182N8Q2 A0A1Y9IVH3 T1PFA4 A0A0L7R869 A0A1A9V182 A0A1B0C6L5 A0A195DC75 A0A1A9X8X2 D3TRK5 A0A195F420 V9IF21 K7J8W7 A0A088AUH9 A0A2A3ERI3 A0A453YNP1 A0A151WQL7 A0A154NXZ8 A0A232EN84 B4Q4I6 B4I1R4 A0A2R5LD00 A0A0J9QWM0 X2J4W8
A0A1B0CSN0 A0A067R3Y8 A0A1L8D952 U5EI74 A0A1L8D9F2 A0A023ELE4 A0A0P6J696 A0A1B6K020 A0A2J7R3A5 A0A336KH59 B0WUC5 A0A1B6MTT8 A0A1Q3FGJ7 A0A1Q3FGJ9 J9JWN4 A0A2P8XV88 T1D4J6 T1E296 A0A2H8TUW5 A0A2S2QJC4 A0A1J1IQF6 A0A069DRJ7 A0A224XID3 T1DLV6 A0A2M3ZG19 W5JM07 A0A1I8JSS7 A0A2M4A3M5 A0A026W2T4 A0A1Q3FGQ1 A0A0V0G5T1 A0A084WD32 A0A034W1F0 A0A0C9RCI8 A0A1Y1KAB1 A0A023F749 N6TWS6 A0A2M4BX00 A0A182JMP6 A0A1W4XFU2 F4WME4 V5GWR6 E0VDR3 A0A158P1X0 A0A1I8JW70 A0A1S4H3B3 A0A1I8JTQ2 A0A182V837 A0A0A1WIU5 A0A0L0CJT7 A0A182KJS3 A0A195BSZ2 Q7PJT0 E2AZJ5 A0A182K0A5 A0A1I8Q3I0 A0A0M8ZW82 E2BHI3 A0A1L8EDW8 A0A195D233 A0A182Q308 R4WDR1 A0A3R7MJV6 A0A1B0A856 A0A182M7T5 A0A0P4WTN1 A0A1L8E940 A0A0K8WK73 A0A0K8UIB5 A0A0K8TQM7 A0A1B6DU58 A0A182N8Q2 A0A1Y9IVH3 T1PFA4 A0A0L7R869 A0A1A9V182 A0A1B0C6L5 A0A195DC75 A0A1A9X8X2 D3TRK5 A0A195F420 V9IF21 K7J8W7 A0A088AUH9 A0A2A3ERI3 A0A453YNP1 A0A151WQL7 A0A154NXZ8 A0A232EN84 B4Q4I6 B4I1R4 A0A2R5LD00 A0A0J9QWM0 X2J4W8
Pubmed
23622113
26354079
22118469
24845553
24945155
26483478
+ More
26999592 29403074 24330624 26334808 20920257 23761445 24508170 30249741 24438588 25348373 28004739 25474469 23537049 21719571 20566863 21347285 12364791 25830018 26108605 20966253 20798317 23691247 26369729 25315136 20353571 20075255 28648823 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867
26999592 29403074 24330624 26334808 20920257 23761445 24508170 30249741 24438588 25348373 28004739 25474469 23537049 21719571 20566863 21347285 12364791 25830018 26108605 20966253 20798317 23691247 26369729 25315136 20353571 20075255 28648823 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867
EMBL
ODYU01010995
SOQ56458.1
RSAL01000024
RVE52216.1
GAIX01006247
JAA86313.1
+ More
KQ459337 KPJ01371.1 AGBW02006209 OWR55234.1 GDQN01006374 GDQN01002465 JAT84680.1 JAT88589.1 AJWK01026310 KK852773 KDR16814.1 GFDF01011106 JAV02978.1 GANO01002740 JAB57131.1 GFDF01011107 JAV02977.1 JXUM01025941 JXUM01025942 JXUM01025943 JXUM01025944 JXUM01025945 GAPW01003552 KQ560739 JAC10046.1 KXJ81061.1 GDUN01000189 JAN95730.1 GECU01002938 JAT04769.1 NEVH01007822 PNF35327.1 UFQS01000300 UFQT01000300 SSX02605.1 SSX22979.1 DS232103 EDS34897.1 GEBQ01000649 JAT39328.1 GFDL01008376 JAV26669.1 GFDL01008374 JAV26671.1 ABLF02023040 PYGN01001295 PSN35929.1 GALA01000933 JAA93919.1 GALA01000934 JAA93918.1 GFXV01006240 MBW18045.1 GGMS01008634 MBY77837.1 CVRI01000054 CRL00737.1 GBGD01002622 JAC86267.1 GFTR01004189 JAW12237.1 GAMD01000221 JAB01370.1 GGFM01006766 MBW27517.1 ADMH02001180 ETN63814.1 GGFK01002038 MBW35359.1 KK107488 QOIP01000012 EZA49901.1 RLU15846.1 GFDL01008321 JAV26724.1 GECL01002771 JAP03353.1 ATLV01022925 KE525338 KFB48126.1 GAKP01009566 JAC49386.1 GBYB01010817 JAG80584.1 GEZM01087881 JAV58422.1 GBBI01001647 JAC17065.1 APGK01052114 KB741209 ENN72846.1 GGFJ01008448 MBW57589.1 GL888217 EGI64788.1 GALX01003758 JAB64708.1 DS235083 EEB11519.1 ADTU01001112 AAAB01008980 APCN01005234 GBXI01015520 JAC98771.1 JRES01000305 KNC32472.1 KQ976418 KYM89475.1 EAA43639.3 GL444248 EFN61128.1 KQ435850 KOX70873.1 GL448301 EFN84836.1 GFDG01001964 JAV16835.1 KQ976948 KYN06932.1 AXCN02001858 AK417764 BAN20979.1 QCYY01000515 ROT84664.1 AXCM01000229 GDRN01003987 JAI67982.1 GFDG01003521 JAV15278.1 GDHF01000763 JAI51551.1 GDHF01026038 GDHF01005596 JAI26276.1 JAI46718.1 GDAI01001137 JAI16466.1 GEDC01008086 JAS29212.1 KA646583 AFP61212.1 KQ414636 KOC67023.1 JXJN01026769 KQ980989 KYN10493.1 CCAG010009992 EZ424057 ADD20333.1 KQ981855 KYN34829.1 JR043067 AEY59718.1 AAZX01000813 KZ288192 PBC34317.1 KQ982831 KYQ50097.1 KQ434778 KZC04432.1 NNAY01003205 OXU19798.1 CM000361 EDX03913.1 CH480820 EDW54471.1 GGLE01003246 MBY07372.1 CM002910 KMY88462.1 AE014134 AHN54178.1
KQ459337 KPJ01371.1 AGBW02006209 OWR55234.1 GDQN01006374 GDQN01002465 JAT84680.1 JAT88589.1 AJWK01026310 KK852773 KDR16814.1 GFDF01011106 JAV02978.1 GANO01002740 JAB57131.1 GFDF01011107 JAV02977.1 JXUM01025941 JXUM01025942 JXUM01025943 JXUM01025944 JXUM01025945 GAPW01003552 KQ560739 JAC10046.1 KXJ81061.1 GDUN01000189 JAN95730.1 GECU01002938 JAT04769.1 NEVH01007822 PNF35327.1 UFQS01000300 UFQT01000300 SSX02605.1 SSX22979.1 DS232103 EDS34897.1 GEBQ01000649 JAT39328.1 GFDL01008376 JAV26669.1 GFDL01008374 JAV26671.1 ABLF02023040 PYGN01001295 PSN35929.1 GALA01000933 JAA93919.1 GALA01000934 JAA93918.1 GFXV01006240 MBW18045.1 GGMS01008634 MBY77837.1 CVRI01000054 CRL00737.1 GBGD01002622 JAC86267.1 GFTR01004189 JAW12237.1 GAMD01000221 JAB01370.1 GGFM01006766 MBW27517.1 ADMH02001180 ETN63814.1 GGFK01002038 MBW35359.1 KK107488 QOIP01000012 EZA49901.1 RLU15846.1 GFDL01008321 JAV26724.1 GECL01002771 JAP03353.1 ATLV01022925 KE525338 KFB48126.1 GAKP01009566 JAC49386.1 GBYB01010817 JAG80584.1 GEZM01087881 JAV58422.1 GBBI01001647 JAC17065.1 APGK01052114 KB741209 ENN72846.1 GGFJ01008448 MBW57589.1 GL888217 EGI64788.1 GALX01003758 JAB64708.1 DS235083 EEB11519.1 ADTU01001112 AAAB01008980 APCN01005234 GBXI01015520 JAC98771.1 JRES01000305 KNC32472.1 KQ976418 KYM89475.1 EAA43639.3 GL444248 EFN61128.1 KQ435850 KOX70873.1 GL448301 EFN84836.1 GFDG01001964 JAV16835.1 KQ976948 KYN06932.1 AXCN02001858 AK417764 BAN20979.1 QCYY01000515 ROT84664.1 AXCM01000229 GDRN01003987 JAI67982.1 GFDG01003521 JAV15278.1 GDHF01000763 JAI51551.1 GDHF01026038 GDHF01005596 JAI26276.1 JAI46718.1 GDAI01001137 JAI16466.1 GEDC01008086 JAS29212.1 KA646583 AFP61212.1 KQ414636 KOC67023.1 JXJN01026769 KQ980989 KYN10493.1 CCAG010009992 EZ424057 ADD20333.1 KQ981855 KYN34829.1 JR043067 AEY59718.1 AAZX01000813 KZ288192 PBC34317.1 KQ982831 KYQ50097.1 KQ434778 KZC04432.1 NNAY01003205 OXU19798.1 CM000361 EDX03913.1 CH480820 EDW54471.1 GGLE01003246 MBY07372.1 CM002910 KMY88462.1 AE014134 AHN54178.1
Proteomes
UP000283053
UP000053268
UP000007151
UP000092461
UP000027135
UP000069940
+ More
UP000249989 UP000235965 UP000002320 UP000007819 UP000245037 UP000183832 UP000000673 UP000069272 UP000053097 UP000279307 UP000030765 UP000019118 UP000075880 UP000192223 UP000007755 UP000009046 UP000005205 UP000076407 UP000075840 UP000075903 UP000037069 UP000075882 UP000078540 UP000007062 UP000000311 UP000075881 UP000095300 UP000053105 UP000008237 UP000078542 UP000075886 UP000283509 UP000092445 UP000075883 UP000075884 UP000075920 UP000095301 UP000053825 UP000078200 UP000092460 UP000078492 UP000092443 UP000092444 UP000078541 UP000002358 UP000005203 UP000242457 UP000075900 UP000075809 UP000076502 UP000215335 UP000000304 UP000001292 UP000000803
UP000249989 UP000235965 UP000002320 UP000007819 UP000245037 UP000183832 UP000000673 UP000069272 UP000053097 UP000279307 UP000030765 UP000019118 UP000075880 UP000192223 UP000007755 UP000009046 UP000005205 UP000076407 UP000075840 UP000075903 UP000037069 UP000075882 UP000078540 UP000007062 UP000000311 UP000075881 UP000095300 UP000053105 UP000008237 UP000078542 UP000075886 UP000283509 UP000092445 UP000075883 UP000075884 UP000075920 UP000095301 UP000053825 UP000078200 UP000092460 UP000078492 UP000092443 UP000092444 UP000078541 UP000002358 UP000005203 UP000242457 UP000075900 UP000075809 UP000076502 UP000215335 UP000000304 UP000001292 UP000000803
PRIDE
Gene 3D
ProteinModelPortal
A0A2H1WTS4
A0A437BP80
S4PAU2
A0A194QD43
A0A212FNB1
A0A1E1WNI1
+ More
A0A1B0CSN0 A0A067R3Y8 A0A1L8D952 U5EI74 A0A1L8D9F2 A0A023ELE4 A0A0P6J696 A0A1B6K020 A0A2J7R3A5 A0A336KH59 B0WUC5 A0A1B6MTT8 A0A1Q3FGJ7 A0A1Q3FGJ9 J9JWN4 A0A2P8XV88 T1D4J6 T1E296 A0A2H8TUW5 A0A2S2QJC4 A0A1J1IQF6 A0A069DRJ7 A0A224XID3 T1DLV6 A0A2M3ZG19 W5JM07 A0A1I8JSS7 A0A2M4A3M5 A0A026W2T4 A0A1Q3FGQ1 A0A0V0G5T1 A0A084WD32 A0A034W1F0 A0A0C9RCI8 A0A1Y1KAB1 A0A023F749 N6TWS6 A0A2M4BX00 A0A182JMP6 A0A1W4XFU2 F4WME4 V5GWR6 E0VDR3 A0A158P1X0 A0A1I8JW70 A0A1S4H3B3 A0A1I8JTQ2 A0A182V837 A0A0A1WIU5 A0A0L0CJT7 A0A182KJS3 A0A195BSZ2 Q7PJT0 E2AZJ5 A0A182K0A5 A0A1I8Q3I0 A0A0M8ZW82 E2BHI3 A0A1L8EDW8 A0A195D233 A0A182Q308 R4WDR1 A0A3R7MJV6 A0A1B0A856 A0A182M7T5 A0A0P4WTN1 A0A1L8E940 A0A0K8WK73 A0A0K8UIB5 A0A0K8TQM7 A0A1B6DU58 A0A182N8Q2 A0A1Y9IVH3 T1PFA4 A0A0L7R869 A0A1A9V182 A0A1B0C6L5 A0A195DC75 A0A1A9X8X2 D3TRK5 A0A195F420 V9IF21 K7J8W7 A0A088AUH9 A0A2A3ERI3 A0A453YNP1 A0A151WQL7 A0A154NXZ8 A0A232EN84 B4Q4I6 B4I1R4 A0A2R5LD00 A0A0J9QWM0 X2J4W8
A0A1B0CSN0 A0A067R3Y8 A0A1L8D952 U5EI74 A0A1L8D9F2 A0A023ELE4 A0A0P6J696 A0A1B6K020 A0A2J7R3A5 A0A336KH59 B0WUC5 A0A1B6MTT8 A0A1Q3FGJ7 A0A1Q3FGJ9 J9JWN4 A0A2P8XV88 T1D4J6 T1E296 A0A2H8TUW5 A0A2S2QJC4 A0A1J1IQF6 A0A069DRJ7 A0A224XID3 T1DLV6 A0A2M3ZG19 W5JM07 A0A1I8JSS7 A0A2M4A3M5 A0A026W2T4 A0A1Q3FGQ1 A0A0V0G5T1 A0A084WD32 A0A034W1F0 A0A0C9RCI8 A0A1Y1KAB1 A0A023F749 N6TWS6 A0A2M4BX00 A0A182JMP6 A0A1W4XFU2 F4WME4 V5GWR6 E0VDR3 A0A158P1X0 A0A1I8JW70 A0A1S4H3B3 A0A1I8JTQ2 A0A182V837 A0A0A1WIU5 A0A0L0CJT7 A0A182KJS3 A0A195BSZ2 Q7PJT0 E2AZJ5 A0A182K0A5 A0A1I8Q3I0 A0A0M8ZW82 E2BHI3 A0A1L8EDW8 A0A195D233 A0A182Q308 R4WDR1 A0A3R7MJV6 A0A1B0A856 A0A182M7T5 A0A0P4WTN1 A0A1L8E940 A0A0K8WK73 A0A0K8UIB5 A0A0K8TQM7 A0A1B6DU58 A0A182N8Q2 A0A1Y9IVH3 T1PFA4 A0A0L7R869 A0A1A9V182 A0A1B0C6L5 A0A195DC75 A0A1A9X8X2 D3TRK5 A0A195F420 V9IF21 K7J8W7 A0A088AUH9 A0A2A3ERI3 A0A453YNP1 A0A151WQL7 A0A154NXZ8 A0A232EN84 B4Q4I6 B4I1R4 A0A2R5LD00 A0A0J9QWM0 X2J4W8
Ontologies
GO
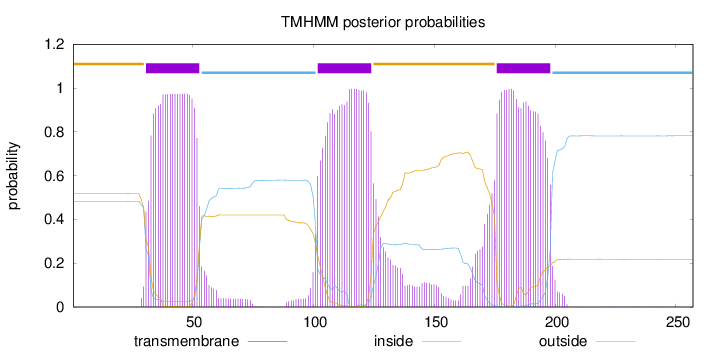
Topology
Length:
257
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
73.7446800000001
Exp number, first 60 AAs:
21.07855
Total prob of N-in:
0.48227
POSSIBLE N-term signal
sequence
outside
1 - 30
TMhelix
31 - 53
inside
54 - 101
TMhelix
102 - 124
outside
125 - 175
TMhelix
176 - 198
inside
199 - 257
Population Genetic Test Statistics
Pi
299.833966
Theta
199.9684
Tajima's D
1.869666
CLR
164.626868
CSRT
0.860806959652017
Interpretation
Uncertain
