Gene
KWMTBOMO08541
Pre Gene Modal
BGIBMGA009321
Annotation
PREDICTED:_probable_asparagine--tRNA_ligase?_mitochondrial_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.303 Mitochondrial Reliability : 1.849
Sequence
CDS
ATGTTGTGTACAAAATTAGAATTTAATAGGTTAAGGAGCTGTAGGCGCTATATTCAAATCAAAGGAATTACTAGAAAATATTGTGCTATTGCAGCTATCGTTGAAAAGTCGGATGTAGGAAGTATTACTGAAGTGAAGGGCTGGGTGAGAAATTTACGTGTCCAAAAAGAGTTTAGTTTTGCTGATATCAGTGATGGTTCGAGCGCGCAGAAATTGCAAATTATAATTCCGAAACATCTGAAAACAGAACAACTAACGTACGGTTCTTCAGTTAAAATTAAAGGAAAACTGTCTCTTAGTCCGCGAGGGCAACTGGAGATTTTAGCTAACGAAGTAAATGTTGTGGGACCTTGCGTCGTCTTAGACGGATATCCATTTAATCCTAGAACTGCACACCCGCCCGAATACACACGCCAATATCTACACTTACGTTCGAGAACGAATTACATATCGGCACTACTCAGAATACGAAGTGCTGTCGTCAAACACATACACGATTTTTTTAATTCAAGAAACTATATCAACATCCACACTCCGATCTTGACTTCAAATGATTGTGAAGGGGCAGGTGAAGTATTTAAAGTTCAACCAGACAACGAGGATACCATAAAAGCAATGATGCAAGAAGGAAAGGATAAAGATTCATTATATTTTGGATTGAAAACATTTCTAACAGTATCTGGTCAGTTGCATTTAGAAGCCATTTGCAGGGGAATGGGTAATGTGTACACACTCGGGCCAACATTTCGAGCAGAAAACTCAAGGTCTAGATTGCATCTCTCTGAGTTTTACATGTTAGAAGCAGAATTAGCATTCTGTGAGAACATTGTCCAGTTGCAAACAGTAATTGAGGAACTTTTAAAATATATATTCACGGAAACAAGAAATACAAATGAGAAAGATATATATTTGATTGATAAGGAAAATAAAAAACCAATTTGGTTGGACAAGCCATTTGTAACTTTGACTTATGATGAAGCTCAGCTTATTTTGAATACAAAGGGGTCTGGAACCGGAGGGGATTTTAATAAAGAACAAGAATTAATTTTGGTAGATCACTGTGGAGGTCCTGTATTTGTAACAAAATGGCCCAAAGACATGAAGTCGTTTTATATGAAAGAAAGCAACAATGACAGTAGTAAGGTTGATGCTTTAGATTTGTTAACACCAGTCACTGGTGAAGTAGTTGGAGGCAGTTTGCGTGAAGACAACTATGAAAAATTAAAACTAAAGCTACCTTCAGAACGACTGCATTGGTACTTAGAATTGCGGAAATTTGGTAACATACCCACAGGAGGATTTGGTTTGGGCTTAGAAAGATTACTACAAGTGCTTTGTGGTGTTAATAATATAAAAGACACACTTCCTTTCCCAAGATGGCCTCATAACTGTGACATGTGA
Protein
MLCTKLEFNRLRSCRRYIQIKGITRKYCAIAAIVEKSDVGSITEVKGWVRNLRVQKEFSFADISDGSSAQKLQIIIPKHLKTEQLTYGSSVKIKGKLSLSPRGQLEILANEVNVVGPCVVLDGYPFNPRTAHPPEYTRQYLHLRSRTNYISALLRIRSAVVKHIHDFFNSRNYINIHTPILTSNDCEGAGEVFKVQPDNEDTIKAMMQEGKDKDSLYFGLKTFLTVSGQLHLEAICRGMGNVYTLGPTFRAENSRSRLHLSEFYMLEAELAFCENIVQLQTVIEELLKYIFTETRNTNEKDIYLIDKENKKPIWLDKPFVTLTYDEAQLILNTKGSGTGGDFNKEQELILVDHCGGPVFVTKWPKDMKSFYMKESNNDSSKVDALDLLTPVTGEVVGGSLREDNYEKLKLKLPSERLHWYLELRKFGNIPTGGFGLGLERLLQVLCGVNNIKDTLPFPRWPHNCDM
Summary
Uniprot
H9JIH1
A0A2H1WTX8
A0A0L7KZM7
A0A0N1IGH0
A0A194Q750
A0A212FNC2
+ More
A0A437BP75 D2A0S8 A0A067R1K2 A0A158NZL0 A0A2P8XV89 A0A154NZK7 A0A2A3ETE9 A0A0L7R8D8 F4WL34 A0A195CI67 A0A0M8ZWA7 A0A151J8X5 A0A026W8X4 A0A3L8D726 A0A088AUH7 A0A195FBI0 A0A232EW07 K7J673 A0A151WM00 A0A195B5D7 A0A1Y1LFX4 A0A1B6DHF1 B4LJC6 E2A438 E2BZI5 A0A0L0BNH6 A0A1B6IVP6 A0A1I8NPC9 B4JVZ4 A0A1Y1LC66 B4KTS7 B3NP36 B4MNG8 A0A1I8NA42 B4HMG0 Q7JZR5 B4P8J0 B4GC55 A0A1W4VBK2 A0A1A9UHG8 W8C1V4 Q290F1 A0A0K8U8V9 B4QII8 A0A182X9I1 A0A182FRN4 A0A182VK96 Q7PX87 A0A1A9Y4H8 D3TPS0 A0A1B0FC45 A0A2M4BNG9 A0A2R7WW59 A0A2M4BNM1 A0A182L8P4 A0A084W0M3 A0A0A1X6X3 A0A3B0J1J0 A0A0M4E3Q7 A0A2M4BM41 A0A182HKG4 A0A182U0I3 A0A182H4W9 A0A182GIN0 A0A0J7KTC3 A0A182K7S2 B3MD79 A0A182IK85 U4U7G8 J3JV90 A0A182PRQ9 W5J5H5 A0A182VVW1 A0A182MMD0 A0A182YBG9 J9JPE3 A0A1Q3F2S3 T1HG00 A0A1B0BDW9 Q16PG0 A0A182QKX2 A0A1J1I2J4 A0A182RA38 A0A0K8TSY2 A0A1B0CNZ0 A0A1A9WIH6 B0W2E8 A0A224XH19 A0A1L8DRI6 A0A023F1W9 A0A1B0DLQ8 A0A224YTG1 A0A146M7L8 A0A131YPK9
A0A437BP75 D2A0S8 A0A067R1K2 A0A158NZL0 A0A2P8XV89 A0A154NZK7 A0A2A3ETE9 A0A0L7R8D8 F4WL34 A0A195CI67 A0A0M8ZWA7 A0A151J8X5 A0A026W8X4 A0A3L8D726 A0A088AUH7 A0A195FBI0 A0A232EW07 K7J673 A0A151WM00 A0A195B5D7 A0A1Y1LFX4 A0A1B6DHF1 B4LJC6 E2A438 E2BZI5 A0A0L0BNH6 A0A1B6IVP6 A0A1I8NPC9 B4JVZ4 A0A1Y1LC66 B4KTS7 B3NP36 B4MNG8 A0A1I8NA42 B4HMG0 Q7JZR5 B4P8J0 B4GC55 A0A1W4VBK2 A0A1A9UHG8 W8C1V4 Q290F1 A0A0K8U8V9 B4QII8 A0A182X9I1 A0A182FRN4 A0A182VK96 Q7PX87 A0A1A9Y4H8 D3TPS0 A0A1B0FC45 A0A2M4BNG9 A0A2R7WW59 A0A2M4BNM1 A0A182L8P4 A0A084W0M3 A0A0A1X6X3 A0A3B0J1J0 A0A0M4E3Q7 A0A2M4BM41 A0A182HKG4 A0A182U0I3 A0A182H4W9 A0A182GIN0 A0A0J7KTC3 A0A182K7S2 B3MD79 A0A182IK85 U4U7G8 J3JV90 A0A182PRQ9 W5J5H5 A0A182VVW1 A0A182MMD0 A0A182YBG9 J9JPE3 A0A1Q3F2S3 T1HG00 A0A1B0BDW9 Q16PG0 A0A182QKX2 A0A1J1I2J4 A0A182RA38 A0A0K8TSY2 A0A1B0CNZ0 A0A1A9WIH6 B0W2E8 A0A224XH19 A0A1L8DRI6 A0A023F1W9 A0A1B0DLQ8 A0A224YTG1 A0A146M7L8 A0A131YPK9
Pubmed
19121390
26227816
26354079
22118469
18362917
19820115
+ More
24845553 21347285 29403074 21719571 24508170 30249741 28648823 20075255 28004739 17994087 20798317 26108605 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 24495485 15632085 22936249 12364791 14747013 17210077 20353571 20966253 24438588 25830018 26483478 23537049 22516182 20920257 23761445 25244985 17510324 26369729 25474469 28797301 26823975 26830274
24845553 21347285 29403074 21719571 24508170 30249741 28648823 20075255 28004739 17994087 20798317 26108605 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 24495485 15632085 22936249 12364791 14747013 17210077 20353571 20966253 24438588 25830018 26483478 23537049 22516182 20920257 23761445 25244985 17510324 26369729 25474469 28797301 26823975 26830274
EMBL
BABH01027222
ODYU01010995
SOQ56457.1
JTDY01004058
KOB68657.1
KQ460817
+ More
KPJ12126.1 KQ459337 KPJ01372.1 AGBW02006209 OWR55235.1 RSAL01000024 RVE52217.1 KQ971338 EFA01641.1 KK852773 KDR16817.1 ADTU01004724 PYGN01001295 PSN35927.1 KQ434778 KZC04428.1 KZ288192 PBC34321.1 KQ414636 KOC67026.1 GL888207 EGI64997.1 KQ977720 KYN00426.1 KQ435850 KOX70869.1 KQ979471 KYN21424.1 KK107353 EZA52086.1 QOIP01000012 RLU16287.1 KQ981685 KYN37985.1 NNAY01001926 OXU22540.1 KQ982944 KYQ48848.1 KQ976587 KYM79723.1 GEZM01061449 JAV70476.1 GEDC01012182 JAS25116.1 CH940648 EDW60506.1 GL436526 EFN71804.1 GL451627 EFN78883.1 JRES01001601 KNC21566.1 GECU01016763 JAS90943.1 CH916375 EDV98132.1 GEZM01061452 JAV70471.1 CH933808 EDW10653.1 CH954179 EDV55675.1 CH963848 EDW73657.1 CH480816 EDW48228.1 AE013599 AY071089 AAF57921.1 AAL48711.1 CM000158 EDW92207.1 CH479181 EDW32398.1 GAMC01000758 JAC05798.1 CM000071 EAL25411.1 GDHF01029217 JAI23097.1 CM000362 CM002911 EDX07435.1 KMY94423.1 AAAB01008987 EAA01739.3 EZ423422 ADD19698.1 CCAG010005008 GGFJ01005403 MBW54544.1 KK855501 PTY22845.1 GGFJ01005400 MBW54541.1 ATLV01019123 KE525262 KFB43767.1 GBXI01007445 GBXI01007117 JAD06847.1 JAD07175.1 OUUW01000001 SPP74807.1 CP012524 ALC40528.1 GGFJ01004870 MBW54011.1 APCN01000828 JXUM01110467 KQ565415 KXJ71003.1 JXUM01066407 KQ562397 KXJ76006.1 LBMM01003414 KMQ93558.1 CH902619 EDV37412.1 KB631829 ERL86566.1 BT127157 AEE62119.1 ADMH02002066 ETN59712.1 AXCM01000480 ABLF02026358 GFDL01013180 JAV21865.1 ACPB03001452 JXJN01012713 CH477784 EAT36240.1 AXCN02001512 CVRI01000036 CRK93070.1 GDAI01000563 JAI17040.1 AJWK01021204 DS231826 EDS28694.1 GFTR01006102 JAW10324.1 GFDF01005011 JAV09073.1 GBBI01003379 JAC15333.1 AJVK01036612 GFPF01006128 MAA17274.1 GDHC01003597 JAQ15032.1 GEDV01007378 JAP81179.1
KPJ12126.1 KQ459337 KPJ01372.1 AGBW02006209 OWR55235.1 RSAL01000024 RVE52217.1 KQ971338 EFA01641.1 KK852773 KDR16817.1 ADTU01004724 PYGN01001295 PSN35927.1 KQ434778 KZC04428.1 KZ288192 PBC34321.1 KQ414636 KOC67026.1 GL888207 EGI64997.1 KQ977720 KYN00426.1 KQ435850 KOX70869.1 KQ979471 KYN21424.1 KK107353 EZA52086.1 QOIP01000012 RLU16287.1 KQ981685 KYN37985.1 NNAY01001926 OXU22540.1 KQ982944 KYQ48848.1 KQ976587 KYM79723.1 GEZM01061449 JAV70476.1 GEDC01012182 JAS25116.1 CH940648 EDW60506.1 GL436526 EFN71804.1 GL451627 EFN78883.1 JRES01001601 KNC21566.1 GECU01016763 JAS90943.1 CH916375 EDV98132.1 GEZM01061452 JAV70471.1 CH933808 EDW10653.1 CH954179 EDV55675.1 CH963848 EDW73657.1 CH480816 EDW48228.1 AE013599 AY071089 AAF57921.1 AAL48711.1 CM000158 EDW92207.1 CH479181 EDW32398.1 GAMC01000758 JAC05798.1 CM000071 EAL25411.1 GDHF01029217 JAI23097.1 CM000362 CM002911 EDX07435.1 KMY94423.1 AAAB01008987 EAA01739.3 EZ423422 ADD19698.1 CCAG010005008 GGFJ01005403 MBW54544.1 KK855501 PTY22845.1 GGFJ01005400 MBW54541.1 ATLV01019123 KE525262 KFB43767.1 GBXI01007445 GBXI01007117 JAD06847.1 JAD07175.1 OUUW01000001 SPP74807.1 CP012524 ALC40528.1 GGFJ01004870 MBW54011.1 APCN01000828 JXUM01110467 KQ565415 KXJ71003.1 JXUM01066407 KQ562397 KXJ76006.1 LBMM01003414 KMQ93558.1 CH902619 EDV37412.1 KB631829 ERL86566.1 BT127157 AEE62119.1 ADMH02002066 ETN59712.1 AXCM01000480 ABLF02026358 GFDL01013180 JAV21865.1 ACPB03001452 JXJN01012713 CH477784 EAT36240.1 AXCN02001512 CVRI01000036 CRK93070.1 GDAI01000563 JAI17040.1 AJWK01021204 DS231826 EDS28694.1 GFTR01006102 JAW10324.1 GFDF01005011 JAV09073.1 GBBI01003379 JAC15333.1 AJVK01036612 GFPF01006128 MAA17274.1 GDHC01003597 JAQ15032.1 GEDV01007378 JAP81179.1
Proteomes
UP000005204
UP000037510
UP000053240
UP000053268
UP000007151
UP000283053
+ More
UP000007266 UP000027135 UP000005205 UP000245037 UP000076502 UP000242457 UP000053825 UP000007755 UP000078542 UP000053105 UP000078492 UP000053097 UP000279307 UP000005203 UP000078541 UP000215335 UP000002358 UP000075809 UP000078540 UP000008792 UP000000311 UP000008237 UP000037069 UP000095300 UP000001070 UP000009192 UP000008711 UP000007798 UP000095301 UP000001292 UP000000803 UP000002282 UP000008744 UP000192221 UP000078200 UP000001819 UP000000304 UP000076407 UP000069272 UP000075903 UP000007062 UP000092443 UP000092444 UP000075882 UP000030765 UP000268350 UP000092553 UP000075840 UP000075902 UP000069940 UP000249989 UP000036403 UP000075881 UP000007801 UP000075880 UP000030742 UP000075885 UP000000673 UP000075920 UP000075883 UP000076408 UP000007819 UP000015103 UP000092460 UP000008820 UP000075886 UP000183832 UP000075900 UP000092461 UP000091820 UP000002320 UP000092462
UP000007266 UP000027135 UP000005205 UP000245037 UP000076502 UP000242457 UP000053825 UP000007755 UP000078542 UP000053105 UP000078492 UP000053097 UP000279307 UP000005203 UP000078541 UP000215335 UP000002358 UP000075809 UP000078540 UP000008792 UP000000311 UP000008237 UP000037069 UP000095300 UP000001070 UP000009192 UP000008711 UP000007798 UP000095301 UP000001292 UP000000803 UP000002282 UP000008744 UP000192221 UP000078200 UP000001819 UP000000304 UP000076407 UP000069272 UP000075903 UP000007062 UP000092443 UP000092444 UP000075882 UP000030765 UP000268350 UP000092553 UP000075840 UP000075902 UP000069940 UP000249989 UP000036403 UP000075881 UP000007801 UP000075880 UP000030742 UP000075885 UP000000673 UP000075920 UP000075883 UP000076408 UP000007819 UP000015103 UP000092460 UP000008820 UP000075886 UP000183832 UP000075900 UP000092461 UP000091820 UP000002320 UP000092462
Pfam
Interpro
IPR012340
NA-bd_OB-fold
+ More
IPR004364 aa-tRNA-synt_II
IPR004365 NA-bd_OB_tRNA
IPR002312 Asp/Asn-tRNA-synth_IIb
IPR004522 Asn-tRNA-ligase
IPR006195 aa-tRNA-synth_II
IPR023865 Aliphatic_acid_kinase_CS
IPR000330 SNF2_N
IPR036306 ISWI_HAND-dom_sf
IPR027417 P-loop_NTPase
IPR038718 SNF2-like_sf
IPR029915 ISWI_metazoa
IPR015195 SLIDE
IPR015194 ISWI_HAND-dom
IPR001005 SANT/Myb
IPR014001 Helicase_ATP-bd
IPR020838 DBINO
IPR001650 Helicase_C
IPR017884 SANT_dom
IPR009057 Homeobox-like_sf
IPR004364 aa-tRNA-synt_II
IPR004365 NA-bd_OB_tRNA
IPR002312 Asp/Asn-tRNA-synth_IIb
IPR004522 Asn-tRNA-ligase
IPR006195 aa-tRNA-synth_II
IPR023865 Aliphatic_acid_kinase_CS
IPR000330 SNF2_N
IPR036306 ISWI_HAND-dom_sf
IPR027417 P-loop_NTPase
IPR038718 SNF2-like_sf
IPR029915 ISWI_metazoa
IPR015195 SLIDE
IPR015194 ISWI_HAND-dom
IPR001005 SANT/Myb
IPR014001 Helicase_ATP-bd
IPR020838 DBINO
IPR001650 Helicase_C
IPR017884 SANT_dom
IPR009057 Homeobox-like_sf
Gene 3D
ProteinModelPortal
H9JIH1
A0A2H1WTX8
A0A0L7KZM7
A0A0N1IGH0
A0A194Q750
A0A212FNC2
+ More
A0A437BP75 D2A0S8 A0A067R1K2 A0A158NZL0 A0A2P8XV89 A0A154NZK7 A0A2A3ETE9 A0A0L7R8D8 F4WL34 A0A195CI67 A0A0M8ZWA7 A0A151J8X5 A0A026W8X4 A0A3L8D726 A0A088AUH7 A0A195FBI0 A0A232EW07 K7J673 A0A151WM00 A0A195B5D7 A0A1Y1LFX4 A0A1B6DHF1 B4LJC6 E2A438 E2BZI5 A0A0L0BNH6 A0A1B6IVP6 A0A1I8NPC9 B4JVZ4 A0A1Y1LC66 B4KTS7 B3NP36 B4MNG8 A0A1I8NA42 B4HMG0 Q7JZR5 B4P8J0 B4GC55 A0A1W4VBK2 A0A1A9UHG8 W8C1V4 Q290F1 A0A0K8U8V9 B4QII8 A0A182X9I1 A0A182FRN4 A0A182VK96 Q7PX87 A0A1A9Y4H8 D3TPS0 A0A1B0FC45 A0A2M4BNG9 A0A2R7WW59 A0A2M4BNM1 A0A182L8P4 A0A084W0M3 A0A0A1X6X3 A0A3B0J1J0 A0A0M4E3Q7 A0A2M4BM41 A0A182HKG4 A0A182U0I3 A0A182H4W9 A0A182GIN0 A0A0J7KTC3 A0A182K7S2 B3MD79 A0A182IK85 U4U7G8 J3JV90 A0A182PRQ9 W5J5H5 A0A182VVW1 A0A182MMD0 A0A182YBG9 J9JPE3 A0A1Q3F2S3 T1HG00 A0A1B0BDW9 Q16PG0 A0A182QKX2 A0A1J1I2J4 A0A182RA38 A0A0K8TSY2 A0A1B0CNZ0 A0A1A9WIH6 B0W2E8 A0A224XH19 A0A1L8DRI6 A0A023F1W9 A0A1B0DLQ8 A0A224YTG1 A0A146M7L8 A0A131YPK9
A0A437BP75 D2A0S8 A0A067R1K2 A0A158NZL0 A0A2P8XV89 A0A154NZK7 A0A2A3ETE9 A0A0L7R8D8 F4WL34 A0A195CI67 A0A0M8ZWA7 A0A151J8X5 A0A026W8X4 A0A3L8D726 A0A088AUH7 A0A195FBI0 A0A232EW07 K7J673 A0A151WM00 A0A195B5D7 A0A1Y1LFX4 A0A1B6DHF1 B4LJC6 E2A438 E2BZI5 A0A0L0BNH6 A0A1B6IVP6 A0A1I8NPC9 B4JVZ4 A0A1Y1LC66 B4KTS7 B3NP36 B4MNG8 A0A1I8NA42 B4HMG0 Q7JZR5 B4P8J0 B4GC55 A0A1W4VBK2 A0A1A9UHG8 W8C1V4 Q290F1 A0A0K8U8V9 B4QII8 A0A182X9I1 A0A182FRN4 A0A182VK96 Q7PX87 A0A1A9Y4H8 D3TPS0 A0A1B0FC45 A0A2M4BNG9 A0A2R7WW59 A0A2M4BNM1 A0A182L8P4 A0A084W0M3 A0A0A1X6X3 A0A3B0J1J0 A0A0M4E3Q7 A0A2M4BM41 A0A182HKG4 A0A182U0I3 A0A182H4W9 A0A182GIN0 A0A0J7KTC3 A0A182K7S2 B3MD79 A0A182IK85 U4U7G8 J3JV90 A0A182PRQ9 W5J5H5 A0A182VVW1 A0A182MMD0 A0A182YBG9 J9JPE3 A0A1Q3F2S3 T1HG00 A0A1B0BDW9 Q16PG0 A0A182QKX2 A0A1J1I2J4 A0A182RA38 A0A0K8TSY2 A0A1B0CNZ0 A0A1A9WIH6 B0W2E8 A0A224XH19 A0A1L8DRI6 A0A023F1W9 A0A1B0DLQ8 A0A224YTG1 A0A146M7L8 A0A131YPK9
PDB
1X56
E-value=3.57283e-54,
Score=536
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
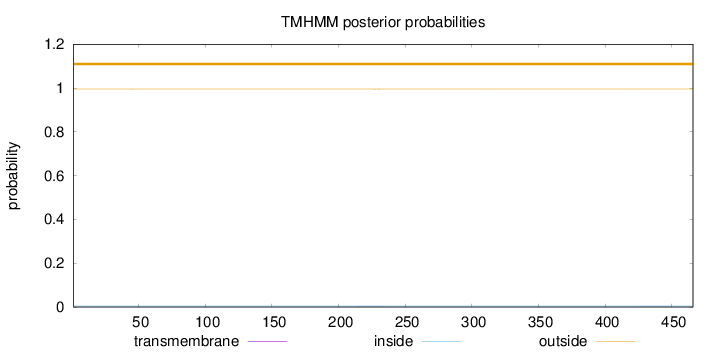
Length:
466
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02026
Exp number, first 60 AAs:
0.00098
Total prob of N-in:
0.00475
outside
1 - 466
Population Genetic Test Statistics
Pi
71.7749
Theta
133.884665
Tajima's D
-0.676471
CLR
112.954747
CSRT
0.206939653017349
Interpretation
Uncertain
