Pre Gene Modal
BGIBMGA009322
Annotation
PREDICTED:_protein_YIPF2_[Papilio_xuthus]
Full name
Protein YIPF
+ More
Probable tRNA N6-adenosine threonylcarbamoyltransferase
Probable tRNA N6-adenosine threonylcarbamoyltransferase
Alternative Name
t(6)A37 threonylcarbamoyladenosine biosynthesis protein
tRNA threonylcarbamoyladenosine biosynthesis protein
tRNA threonylcarbamoyladenosine biosynthesis protein
Location in the cell
PlasmaMembrane Reliability : 4.71
Sequence
CDS
ATGTCGAACGTCCATACGGGCGAGTTGTTAAATTTCCAGGACTATTCCCCGGAACACAATGAGCGAACTGCGCGTATTGATGTAGAGGCAGTACATACAAATCAATACAATAATAGTAATATTTCTAATCAAAATTACTCCTTTGACGAAACGCCCGGAGCTGTTCCAGAAGCAGAGACTCAACCTCAATCGAATCACAACTTCTGGACAATAGAGTACTATCAGAAATACTTTGATGTGCAGACCAGTGAAGTTGTGGAGAGGATTATATCATCAGTATTGCCCCAGAAAGTCTCTAGAAACTATTTTGATGAGAAGATAAAAGGCAAACCCGACTTGTATGGACCTATCTGGATTTCATTGACTCTGATCTTCACAATTGCCGTGAGCGGGAACATAGCAAGTTATCTACAGAATGTTAACAAGGCAGTCCACTGGCGCTATGATTTTCATTTGGTGTCTTACGCAGCAACAGCCATAGTATGTTACGTTTGGTTGGTCCCTCTGGCACTGTGGGGTGTCTTGAAATGGACTACGCTTCCCGATGGACAAGATGAGATTGAGACGCAGACCTCAAACTCTCCAACTATGATATCTCTCTTCTGTTTATATGGCTATTCGCTTTCCATTTACATCCCTGTGGCTATATTCTGGACCATCCAGATCTCTTGGCTCCAGTGGTTGTTTGTGTTGATGGCTGCCTTTGTGTCCGGAGCAGTCCTCATATTCTGGCTGCTGCCAGCTTTGAAGAAATCAAAATATTTTTATATACTTGTCGGATCTATTCTTGCTTTTCATTTCTTGTTGGCAACTGGTTTTATGTTATACTTCTTTCACGGACCAAACGCACATGACTAA
Protein
MSNVHTGELLNFQDYSPEHNERTARIDVEAVHTNQYNNSNISNQNYSFDETPGAVPEAETQPQSNHNFWTIEYYQKYFDVQTSEVVERIISSVLPQKVSRNYFDEKIKGKPDLYGPIWISLTLIFTIAVSGNIASYLQNVNKAVHWRYDFHLVSYAATAIVCYVWLVPLALWGVLKWTTLPDGQDEIETQTSNSPTMISLFCLYGYSLSIYIPVAIFWTIQISWLQWLFVLMAAFVSGAVLIFWLLPALKKSKYFYILVGSILAFHFLLATGFMLYFFHGPNAHD
Summary
Description
Component of the EKC/KEOPS complex that is required for the formation of a threonylcarbamoyl group on adenosine at position 37 (t(6)A37) in tRNAs that read codons beginning with adenine. The complex is probably involved in the transfer of the threonylcarbamoyl moiety of threonylcarbamoyl-AMP (TC-AMP) to the N6 group of A37. Likely plays a direct catalytic role in this reaction, but requires other protein(s) of the complex to fulfill this activity.
Catalytic Activity
adenosine(37) in tRNA + L-threonylcarbamoyladenylate = AMP + H(+) + N(6)-L-threonylcarbamoyladenosine(37) in tRNA
Cofactor
a divalent metal cation
Subunit
Component of the EKC/KEOPS complex; the whole complex dimerizes.
Similarity
Belongs to the YIP1 family.
Belongs to the KAE1 / TsaD family.
Belongs to the KAE1 / TsaD family.
Uniprot
H9JIH2
A0A194Q7K8
A0A212FNC7
A0A1E1W2L8
A0A437BPI1
A0A2H1WTQ9
+ More
A0A0L7LLL3 D2A1I7 A0A1L8D906 A0A1B0EUT3 N6TWP7 J3JXS0 A0A0K8TQQ8 A0A1Y1LWU4 A0A034W9N0 A0A1I8JUP4 A0A088AV47 A0A2A3ESL8 W5JT29 A0A0K8VAS3 A0A182Y2C7 A0A0A1WEQ1 A0A2M3ZGX7 A0A2M3Z3K9 A0A0J7K9B8 A0A1L8E8A9 A0A1L8E7Q2 E1ZZ84 A0A1L8E8E4 A0A1L8ED40 A0A2M4AC14 A0A2M4ABJ0 A0A2M4ABC9 A0A1I8ML86 A0A2M4ABT8 T1PJC1 A0A154NXJ2 A0A1W4XTR1 A0A182MTJ3 A0A1I8NY11 A0A182QGV3 A0A0M8ZU17 A0A2P8XV93 B4L603 B4M750 A0A0L0C396 A0A1A9WWX0 E9J1A9 A0A182S7F9 A0A2M4BUL5 A0A2M4BUQ2 A0A0M3QZ01 A0A2M4BUN7 A0A023EQ34 A0A023EMS2 A0A026W3P5 B4JM02 A0A182J7G1 A0A0C9RY25 A0A1J1IXL2 A0A3B0KZ32 Q17KU7 B3MS24 B4IGK6 A0A182UW57 A0A1A9VWP9 Q29FT9 B4NEI2 Q7QIM4 B4Q1Y9 A0A182JP76 A0A453YK63 A0A182UIG1 B3NX85 Q9VYI1 A0A1W4VL09 A0A182PII2 A0A084WEZ4 A0A1B0G811 A0A067R3Z1 A0A2J7R3C6 K7J5P4 A0A182WWJ0 A0A1B6DHK5 A0A182L3M2 A0A232FJ33 F4X8R3 A0A195FCS3 A0A195DSE0 A0A1B6ME12 E0V988 A0A182WBJ2 A0A195B9L7 A0A158NBG3 A0A1B6JCZ7 A0A0L7R7P9 W8CB19
A0A0L7LLL3 D2A1I7 A0A1L8D906 A0A1B0EUT3 N6TWP7 J3JXS0 A0A0K8TQQ8 A0A1Y1LWU4 A0A034W9N0 A0A1I8JUP4 A0A088AV47 A0A2A3ESL8 W5JT29 A0A0K8VAS3 A0A182Y2C7 A0A0A1WEQ1 A0A2M3ZGX7 A0A2M3Z3K9 A0A0J7K9B8 A0A1L8E8A9 A0A1L8E7Q2 E1ZZ84 A0A1L8E8E4 A0A1L8ED40 A0A2M4AC14 A0A2M4ABJ0 A0A2M4ABC9 A0A1I8ML86 A0A2M4ABT8 T1PJC1 A0A154NXJ2 A0A1W4XTR1 A0A182MTJ3 A0A1I8NY11 A0A182QGV3 A0A0M8ZU17 A0A2P8XV93 B4L603 B4M750 A0A0L0C396 A0A1A9WWX0 E9J1A9 A0A182S7F9 A0A2M4BUL5 A0A2M4BUQ2 A0A0M3QZ01 A0A2M4BUN7 A0A023EQ34 A0A023EMS2 A0A026W3P5 B4JM02 A0A182J7G1 A0A0C9RY25 A0A1J1IXL2 A0A3B0KZ32 Q17KU7 B3MS24 B4IGK6 A0A182UW57 A0A1A9VWP9 Q29FT9 B4NEI2 Q7QIM4 B4Q1Y9 A0A182JP76 A0A453YK63 A0A182UIG1 B3NX85 Q9VYI1 A0A1W4VL09 A0A182PII2 A0A084WEZ4 A0A1B0G811 A0A067R3Z1 A0A2J7R3C6 K7J5P4 A0A182WWJ0 A0A1B6DHK5 A0A182L3M2 A0A232FJ33 F4X8R3 A0A195FCS3 A0A195DSE0 A0A1B6ME12 E0V988 A0A182WBJ2 A0A195B9L7 A0A158NBG3 A0A1B6JCZ7 A0A0L7R7P9 W8CB19
EC Number
2.3.1.234
Pubmed
19121390
26354079
22118469
26227816
18362917
19820115
+ More
23537049 22516182 26369729 28004739 25348373 20920257 23761445 25244985 25830018 20798317 25315136 29403074 17994087 26108605 21282665 24945155 26483478 24508170 30249741 17510324 15632085 12364791 14747013 17210077 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24438588 24845553 20075255 20966253 28648823 21719571 20566863 21347285 24495485
23537049 22516182 26369729 28004739 25348373 20920257 23761445 25244985 25830018 20798317 25315136 29403074 17994087 26108605 21282665 24945155 26483478 24508170 30249741 17510324 15632085 12364791 14747013 17210077 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24438588 24845553 20075255 20966253 28648823 21719571 20566863 21347285 24495485
EMBL
BABH01027219
KQ459337
KPJ01374.1
AGBW02006209
OWR55237.1
GDQN01009831
+ More
JAT81223.1 RSAL01000023 RVE52292.1 ODYU01010995 SOQ56455.1 JTDY01000649 KOB76337.1 KQ971338 EFA01533.2 GFDF01011143 JAV02941.1 AJWK01034750 APGK01002895 APGK01058069 KB741286 KB735013 ENN70727.1 ENN83468.1 APGK01058067 BT128041 KB632373 AEE63002.1 ENN70725.1 ERL93792.1 GDAI01001115 JAI16488.1 GEZM01044978 JAV77999.1 GAKP01008459 JAC50493.1 KZ288192 PBC34272.1 ADMH02000528 ETN66095.1 GDHF01016368 JAI35946.1 GBXI01017121 JAC97170.1 GGFM01006984 MBW27735.1 GGFM01002358 MBW23109.1 LBMM01011370 KMQ86884.1 GFDG01003987 JAV14812.1 GFDG01004039 JAV14760.1 GL435311 EFN73499.1 GFDG01003943 JAV14856.1 GFDG01002235 JAV16564.1 GGFK01004941 MBW38262.1 GGFK01004834 MBW38155.1 GGFK01004774 MBW38095.1 GGFK01004926 MBW38247.1 KA648003 AFP62632.1 KQ434778 KZC04379.1 AXCM01000934 AXCN02000558 KQ435850 KOX70937.1 PYGN01001295 PSN35932.1 CH933812 EDW05799.1 CH940653 EDW62617.1 JRES01000955 KNC26805.1 GL767586 EFZ13413.1 GGFJ01007563 MBW56704.1 GGFJ01007581 MBW56722.1 CP012528 ALC48509.1 GGFJ01007582 MBW56723.1 GAPW01002939 JAC10659.1 JXUM01020992 JXUM01037730 GAPW01002940 KQ561140 KQ560588 JAC10658.1 KXJ79593.1 KXJ81769.1 KK107453 QOIP01000004 EZA50618.1 RLU24114.1 CH916371 EDV91763.1 GBYB01014175 JAG83942.1 CVRI01000060 CRL03902.1 OUUW01000021 SPP89388.1 CH477221 EAT47307.1 CH902622 EDV34579.1 CH480836 EDW48956.1 CH379065 EAL31490.1 CH964239 EDW82151.1 AAAB01008807 EAA04402.4 CM000162 EDX01510.1 APCN01003693 CH954180 EDV47257.1 AE014298 AY058677 AAF48214.2 AAL13906.1 ATLV01023266 KE525341 KFB48788.1 CCAG010000152 KK852773 KDR16819.1 NEVH01007822 PNF35326.1 GEDC01012198 GEDC01009944 JAS25100.1 JAS27354.1 NNAY01000131 OXU30685.1 GL888932 EGI57332.1 KQ981673 KYN38208.1 KQ980581 KYN15434.1 GEBQ01005815 JAT34162.1 DS234991 EEB09944.1 KQ976540 KYM81223.1 ADTU01011059 GECU01010709 JAS96997.1 KQ414638 KOC66900.1 GAMC01005226 JAC01330.1
JAT81223.1 RSAL01000023 RVE52292.1 ODYU01010995 SOQ56455.1 JTDY01000649 KOB76337.1 KQ971338 EFA01533.2 GFDF01011143 JAV02941.1 AJWK01034750 APGK01002895 APGK01058069 KB741286 KB735013 ENN70727.1 ENN83468.1 APGK01058067 BT128041 KB632373 AEE63002.1 ENN70725.1 ERL93792.1 GDAI01001115 JAI16488.1 GEZM01044978 JAV77999.1 GAKP01008459 JAC50493.1 KZ288192 PBC34272.1 ADMH02000528 ETN66095.1 GDHF01016368 JAI35946.1 GBXI01017121 JAC97170.1 GGFM01006984 MBW27735.1 GGFM01002358 MBW23109.1 LBMM01011370 KMQ86884.1 GFDG01003987 JAV14812.1 GFDG01004039 JAV14760.1 GL435311 EFN73499.1 GFDG01003943 JAV14856.1 GFDG01002235 JAV16564.1 GGFK01004941 MBW38262.1 GGFK01004834 MBW38155.1 GGFK01004774 MBW38095.1 GGFK01004926 MBW38247.1 KA648003 AFP62632.1 KQ434778 KZC04379.1 AXCM01000934 AXCN02000558 KQ435850 KOX70937.1 PYGN01001295 PSN35932.1 CH933812 EDW05799.1 CH940653 EDW62617.1 JRES01000955 KNC26805.1 GL767586 EFZ13413.1 GGFJ01007563 MBW56704.1 GGFJ01007581 MBW56722.1 CP012528 ALC48509.1 GGFJ01007582 MBW56723.1 GAPW01002939 JAC10659.1 JXUM01020992 JXUM01037730 GAPW01002940 KQ561140 KQ560588 JAC10658.1 KXJ79593.1 KXJ81769.1 KK107453 QOIP01000004 EZA50618.1 RLU24114.1 CH916371 EDV91763.1 GBYB01014175 JAG83942.1 CVRI01000060 CRL03902.1 OUUW01000021 SPP89388.1 CH477221 EAT47307.1 CH902622 EDV34579.1 CH480836 EDW48956.1 CH379065 EAL31490.1 CH964239 EDW82151.1 AAAB01008807 EAA04402.4 CM000162 EDX01510.1 APCN01003693 CH954180 EDV47257.1 AE014298 AY058677 AAF48214.2 AAL13906.1 ATLV01023266 KE525341 KFB48788.1 CCAG010000152 KK852773 KDR16819.1 NEVH01007822 PNF35326.1 GEDC01012198 GEDC01009944 JAS25100.1 JAS27354.1 NNAY01000131 OXU30685.1 GL888932 EGI57332.1 KQ981673 KYN38208.1 KQ980581 KYN15434.1 GEBQ01005815 JAT34162.1 DS234991 EEB09944.1 KQ976540 KYM81223.1 ADTU01011059 GECU01010709 JAS96997.1 KQ414638 KOC66900.1 GAMC01005226 JAC01330.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000283053
UP000037510
UP000007266
+ More
UP000092461 UP000019118 UP000030742 UP000075900 UP000005203 UP000242457 UP000000673 UP000076408 UP000036403 UP000000311 UP000095301 UP000076502 UP000192223 UP000075883 UP000095300 UP000075886 UP000053105 UP000245037 UP000009192 UP000008792 UP000037069 UP000091820 UP000075901 UP000092553 UP000069940 UP000249989 UP000053097 UP000279307 UP000001070 UP000075880 UP000183832 UP000268350 UP000008820 UP000007801 UP000001292 UP000075903 UP000078200 UP000001819 UP000007798 UP000007062 UP000002282 UP000075881 UP000075840 UP000075902 UP000008711 UP000000803 UP000192221 UP000075885 UP000030765 UP000092444 UP000027135 UP000235965 UP000002358 UP000076407 UP000075882 UP000215335 UP000007755 UP000078541 UP000078492 UP000009046 UP000075920 UP000078540 UP000005205 UP000053825
UP000092461 UP000019118 UP000030742 UP000075900 UP000005203 UP000242457 UP000000673 UP000076408 UP000036403 UP000000311 UP000095301 UP000076502 UP000192223 UP000075883 UP000095300 UP000075886 UP000053105 UP000245037 UP000009192 UP000008792 UP000037069 UP000091820 UP000075901 UP000092553 UP000069940 UP000249989 UP000053097 UP000279307 UP000001070 UP000075880 UP000183832 UP000268350 UP000008820 UP000007801 UP000001292 UP000075903 UP000078200 UP000001819 UP000007798 UP000007062 UP000002282 UP000075881 UP000075840 UP000075902 UP000008711 UP000000803 UP000192221 UP000075885 UP000030765 UP000092444 UP000027135 UP000235965 UP000002358 UP000076407 UP000075882 UP000215335 UP000007755 UP000078541 UP000078492 UP000009046 UP000075920 UP000078540 UP000005205 UP000053825
Interpro
SUPFAM
SSF48264
SSF48264
Gene 3D
ProteinModelPortal
H9JIH2
A0A194Q7K8
A0A212FNC7
A0A1E1W2L8
A0A437BPI1
A0A2H1WTQ9
+ More
A0A0L7LLL3 D2A1I7 A0A1L8D906 A0A1B0EUT3 N6TWP7 J3JXS0 A0A0K8TQQ8 A0A1Y1LWU4 A0A034W9N0 A0A1I8JUP4 A0A088AV47 A0A2A3ESL8 W5JT29 A0A0K8VAS3 A0A182Y2C7 A0A0A1WEQ1 A0A2M3ZGX7 A0A2M3Z3K9 A0A0J7K9B8 A0A1L8E8A9 A0A1L8E7Q2 E1ZZ84 A0A1L8E8E4 A0A1L8ED40 A0A2M4AC14 A0A2M4ABJ0 A0A2M4ABC9 A0A1I8ML86 A0A2M4ABT8 T1PJC1 A0A154NXJ2 A0A1W4XTR1 A0A182MTJ3 A0A1I8NY11 A0A182QGV3 A0A0M8ZU17 A0A2P8XV93 B4L603 B4M750 A0A0L0C396 A0A1A9WWX0 E9J1A9 A0A182S7F9 A0A2M4BUL5 A0A2M4BUQ2 A0A0M3QZ01 A0A2M4BUN7 A0A023EQ34 A0A023EMS2 A0A026W3P5 B4JM02 A0A182J7G1 A0A0C9RY25 A0A1J1IXL2 A0A3B0KZ32 Q17KU7 B3MS24 B4IGK6 A0A182UW57 A0A1A9VWP9 Q29FT9 B4NEI2 Q7QIM4 B4Q1Y9 A0A182JP76 A0A453YK63 A0A182UIG1 B3NX85 Q9VYI1 A0A1W4VL09 A0A182PII2 A0A084WEZ4 A0A1B0G811 A0A067R3Z1 A0A2J7R3C6 K7J5P4 A0A182WWJ0 A0A1B6DHK5 A0A182L3M2 A0A232FJ33 F4X8R3 A0A195FCS3 A0A195DSE0 A0A1B6ME12 E0V988 A0A182WBJ2 A0A195B9L7 A0A158NBG3 A0A1B6JCZ7 A0A0L7R7P9 W8CB19
A0A0L7LLL3 D2A1I7 A0A1L8D906 A0A1B0EUT3 N6TWP7 J3JXS0 A0A0K8TQQ8 A0A1Y1LWU4 A0A034W9N0 A0A1I8JUP4 A0A088AV47 A0A2A3ESL8 W5JT29 A0A0K8VAS3 A0A182Y2C7 A0A0A1WEQ1 A0A2M3ZGX7 A0A2M3Z3K9 A0A0J7K9B8 A0A1L8E8A9 A0A1L8E7Q2 E1ZZ84 A0A1L8E8E4 A0A1L8ED40 A0A2M4AC14 A0A2M4ABJ0 A0A2M4ABC9 A0A1I8ML86 A0A2M4ABT8 T1PJC1 A0A154NXJ2 A0A1W4XTR1 A0A182MTJ3 A0A1I8NY11 A0A182QGV3 A0A0M8ZU17 A0A2P8XV93 B4L603 B4M750 A0A0L0C396 A0A1A9WWX0 E9J1A9 A0A182S7F9 A0A2M4BUL5 A0A2M4BUQ2 A0A0M3QZ01 A0A2M4BUN7 A0A023EQ34 A0A023EMS2 A0A026W3P5 B4JM02 A0A182J7G1 A0A0C9RY25 A0A1J1IXL2 A0A3B0KZ32 Q17KU7 B3MS24 B4IGK6 A0A182UW57 A0A1A9VWP9 Q29FT9 B4NEI2 Q7QIM4 B4Q1Y9 A0A182JP76 A0A453YK63 A0A182UIG1 B3NX85 Q9VYI1 A0A1W4VL09 A0A182PII2 A0A084WEZ4 A0A1B0G811 A0A067R3Z1 A0A2J7R3C6 K7J5P4 A0A182WWJ0 A0A1B6DHK5 A0A182L3M2 A0A232FJ33 F4X8R3 A0A195FCS3 A0A195DSE0 A0A1B6ME12 E0V988 A0A182WBJ2 A0A195B9L7 A0A158NBG3 A0A1B6JCZ7 A0A0L7R7P9 W8CB19
Ontologies
GO
PANTHER
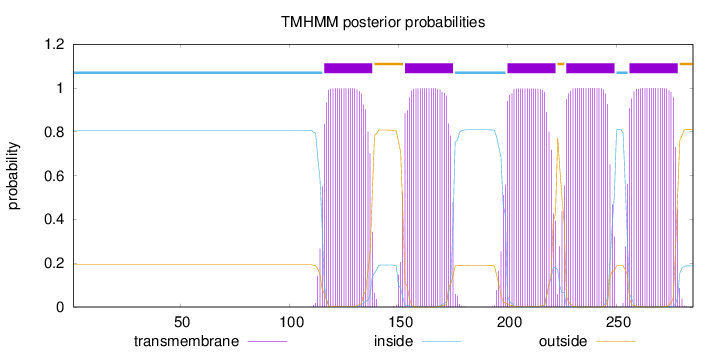
Topology
Subcellular location
Membrane
Cytoplasm
Nucleus
Cytoplasm
Nucleus
Length:
285
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
113.31461
Exp number, first 60 AAs:
0
Total prob of N-in:
0.80723
inside
1 - 115
TMhelix
116 - 138
outside
139 - 152
TMhelix
153 - 175
inside
176 - 199
TMhelix
200 - 222
outside
223 - 226
TMhelix
227 - 249
inside
250 - 255
TMhelix
256 - 278
outside
279 - 285
Population Genetic Test Statistics
Pi
408.331653
Theta
211.38471
Tajima's D
2.996675
CLR
0.168174
CSRT
0.979801009949502
Interpretation
Uncertain
