Gene
KWMTBOMO08537 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009323
Annotation
PREDICTED:_putative_oxidoreductase_GLYR1_homolog_[Bombyx_mori]
Full name
Putative oxidoreductase GLYR1 homolog
Alternative Name
Glyoxylate reductase 1 homolog
Nuclear protein NP60 homolog
Nuclear protein NP60 homolog
Location in the cell
Nuclear Reliability : 2.409
Sequence
CDS
ATGTCTGAAGGATATAAACTTGGGGATTTAGTTTGGGCGAAAATGAAGGGTTTCAGTCCTTGGCCTGGCCGGGTAGCGGTTCCCACTCCTGAACTTAGAACGCCGAAGAAGGCCACCAACGTACAATGCATTTACTTCTTTGGTACAAATAACTATGCTTGGATCGAAGACATCAACATCAAGCCGTACCAAGAGTTTAAGGAGCAGTTGATCAAATCCTGCAAGACGGCTGCGTTTAAAGAAGCCGTGAATCAAATCGAAGACTTCATTGAACATCCTGAGAAATTCGGAGATCTAGACGACCACACCCGTAACGTGGAGGAGGAATTCGATAAACTGAAGGAGAACGTCGAAGGAGACGACAGCAAAGAGGAGGCGACTGAAGCTGATGATGCACCCGCTCCAGAGTCTGCGAAGAAAAAGAGTTCCACGCCTAAGATACGTTTGGGCATTTCAAAGGTGAAGCCTATAAAGCGTGCCTCGTCGGCCGGCGTCAAGTCCACGCCGCGGAAGAAGGCAAAGAGTTCCCTCTCCAGGTCGTACACCGACTCCGATGTCGCCAACGAGAAGCCGTCGATGCTGAACCACTCGTTCTCCAAGAAGTCCAGCCTGTTGCAACGTCCCTCGAATATATTGAGACCTGAGACTCCCCCGCTGGACCTGGAGAACGTTTCTGAGACTCTTCTAGAGAAGAACATCAAGGCCAGCCAGATGAAGTTCGGCTTCCTCGGACTCGGCATCATGGGCTCGGGTATCGTGAAGAACCTCCTCAACTCGGGACACAAGGTCATCGTTTGGAACAGAACCGCGGCCAAGAAATATCTTCAGGCTCTTGTCTAA
Protein
MSEGYKLGDLVWAKMKGFSPWPGRVAVPTPELRTPKKATNVQCIYFFGTNNYAWIEDINIKPYQEFKEQLIKSCKTAAFKEAVNQIEDFIEHPEKFGDLDDHTRNVEEEFDKLKENVEGDDSKEEATEADDAPAPESAKKKSSTPKIRLGISKVKPIKRASSAGVKSTPRKKAKSSLSRSYTDSDVANEKPSMLNHSFSKKSSLLQRPSNILRPETPPLDLENVSETLLEKNIKASQMKFGFLGLGIMGSGIVKNLLNSGHKVIVWNRTAAKKYLQALV
Summary
Description
May have oxidoreductase activity.
Miscellaneous
The conserved NAD-binding sites suggest that this protein may have oxidoreductase activity.
Similarity
Belongs to the HIBADH-related family. NP60 subfamily.
Keywords
Complete proteome
NAD
Oxidoreductase
Reference proteome
Feature
chain Putative oxidoreductase GLYR1 homolog
Uniprot
A0A2H1WTS1
A0A3S2M6T9
A0A2A4J6Z2
A0A212FNF2
A0A2A4J7W9
A0A0N0PBT8
+ More
H9JIH3 A0A194QD48 A0A151X995 A0A158P1G7 E2A676 A0A3L8D5T8 E2BGS4 A0A088AUI8 K7J4H0 A0A2J7PGC3 A0A2A3ERD7 V9IAL3 A0A1B6DF01 A0A1B6CNC1 A0A182HAH3 A0A0C9QUR3 A0A1Q3G210 A0A084VWE3 A0A1B6CWT0 A0A182IWZ4 A0A182LYP8 A0A0P6JJ89 A0A0P5Q8I8 A0A0P4ZBF1 A0A182RRC7 A0A0P5XKV4 A0A1S4FEC3 A0A0P4YIZ8 A0A1B6HZ81 A0A0P4WQ01 A0A182W8V8 A0A0P5B673 A0A182Y6D8 Q175F8 F4W9B6 A0A0L7RKI4 A0A1B6GI18 A0A0N0U415 A0A1B0D1N2 A0A182QM25 A0A182NAW7 A0A182K4N2 A0A2M4A6P4 A0A182HRD4 A0A453YVI5 A0A182PNH1 A0A1S4H2H3 A0A182KZ01 A0A182VBT9 A0A1L8DAW2 W5J4J7 A0A2M3Z2I2 A0A2M4BIM9 A0A182F4C5 A0A182XHU2 Q7Q161 E0VKD9 A0A1Y1LFR7 A0A182TI87 A0A0P6BML2 A0A195CIC9 A0A0K8TN33 A0A0P5TIF6 A0A195F2Q6 U5EZK8 A0A151J3V6 A0A026W1G1 A0A1W4WU51 A0A336LMM2 J9JU51 A0A0P5HI90 A0A154NXI6 A0A0P5EAW3 A0A2R5L8C0 A0A1Z5L400 A0A0P5QGV7 A0A0P4WCV3 A0A224YLT0 A0A131YTV8 A0A195BRB1 A0A2S2PMH5 T1HKM5 A0A2P8Y0H3 E9H750 A0A1D2N4N3 A0A444SCH0 A0A1J1IN33 A0A1S4FWH1 D2A023 A0A1I8Q3G3 A0A1I8Q3G5 A0A1I8Q3E7
H9JIH3 A0A194QD48 A0A151X995 A0A158P1G7 E2A676 A0A3L8D5T8 E2BGS4 A0A088AUI8 K7J4H0 A0A2J7PGC3 A0A2A3ERD7 V9IAL3 A0A1B6DF01 A0A1B6CNC1 A0A182HAH3 A0A0C9QUR3 A0A1Q3G210 A0A084VWE3 A0A1B6CWT0 A0A182IWZ4 A0A182LYP8 A0A0P6JJ89 A0A0P5Q8I8 A0A0P4ZBF1 A0A182RRC7 A0A0P5XKV4 A0A1S4FEC3 A0A0P4YIZ8 A0A1B6HZ81 A0A0P4WQ01 A0A182W8V8 A0A0P5B673 A0A182Y6D8 Q175F8 F4W9B6 A0A0L7RKI4 A0A1B6GI18 A0A0N0U415 A0A1B0D1N2 A0A182QM25 A0A182NAW7 A0A182K4N2 A0A2M4A6P4 A0A182HRD4 A0A453YVI5 A0A182PNH1 A0A1S4H2H3 A0A182KZ01 A0A182VBT9 A0A1L8DAW2 W5J4J7 A0A2M3Z2I2 A0A2M4BIM9 A0A182F4C5 A0A182XHU2 Q7Q161 E0VKD9 A0A1Y1LFR7 A0A182TI87 A0A0P6BML2 A0A195CIC9 A0A0K8TN33 A0A0P5TIF6 A0A195F2Q6 U5EZK8 A0A151J3V6 A0A026W1G1 A0A1W4WU51 A0A336LMM2 J9JU51 A0A0P5HI90 A0A154NXI6 A0A0P5EAW3 A0A2R5L8C0 A0A1Z5L400 A0A0P5QGV7 A0A0P4WCV3 A0A224YLT0 A0A131YTV8 A0A195BRB1 A0A2S2PMH5 T1HKM5 A0A2P8Y0H3 E9H750 A0A1D2N4N3 A0A444SCH0 A0A1J1IN33 A0A1S4FWH1 D2A023 A0A1I8Q3G3 A0A1I8Q3G5 A0A1I8Q3E7
EC Number
1.-.-.-
Pubmed
EMBL
ODYU01010995
SOQ56453.1
RSAL01000023
RVE52294.1
NWSH01002610
PCG67855.1
+ More
AGBW02006209 OWR55239.1 PCG67856.1 KQ460817 KPJ12129.1 BABH01027215 KQ459337 KPJ01376.1 KQ982402 KYQ56858.1 ADTU01001056 ADTU01001057 ADTU01001058 ADTU01001059 ADTU01001060 ADTU01001061 GL437116 EFN71056.1 QOIP01000012 RLU15847.1 GL448228 EFN85056.1 NEVH01025635 PNF15379.1 KZ288192 PBC34298.1 JR038208 AEY58158.1 GEDC01013039 JAS24259.1 GEDC01022319 JAS14979.1 JXUM01031210 KQ560913 KXJ80367.1 GBYB01007492 GBYB01007494 JAG77259.1 JAG77261.1 GFDL01001212 JAV33833.1 ATLV01017581 KE525174 KFB42287.1 GEDC01019368 JAS17930.1 AXCM01002228 GDIQ01005978 JAN88759.1 GDIQ01117697 JAL34029.1 GDIP01228083 JAI95318.1 GDIP01070605 JAM33110.1 GDIP01228084 JAI95317.1 GECU01027695 JAS80011.1 GDIP01252332 LRGB01000872 JAI71069.1 KZS15433.1 GDIP01188990 JAJ34412.1 CH477401 GL888002 EGI69298.1 KQ414572 KOC71328.1 GECZ01007685 JAS62084.1 KQ435850 KOX70902.1 AJVK01010334 AJVK01010335 AXCN02000653 GGFK01003089 MBW36410.1 APCN01001690 AAAB01008980 GFDF01010486 JAV03598.1 ADMH02002138 ETN58343.1 GGFM01001964 MBW22715.1 GGFJ01003778 MBW52919.1 DS235243 EEB13855.1 GEZM01057056 JAV72474.1 GDIP01012526 JAM91189.1 KQ977720 KYN00496.1 GDAI01002062 JAI15541.1 GDIP01126305 JAL77409.1 KQ981855 KYN34855.1 GANO01000613 JAB59258.1 KQ980275 KYN17007.1 KK107488 EZA49910.1 UFQT01000023 SSX18021.1 ABLF02017887 ABLF02017889 ABLF02062618 GDIQ01227108 JAK24617.1 KQ434778 KZC04369.1 GDIP01144090 JAJ79312.1 GGLE01001541 MBY05667.1 GFJQ02004891 JAW02079.1 GDIQ01124086 JAL27640.1 GDRN01067921 JAI64295.1 GFPF01005553 MAA16699.1 GEDV01006170 JAP82387.1 KQ976418 KYM89496.1 GGMR01018042 MBY30661.1 ACPB03006490 PYGN01001092 PSN37674.1 GL732599 EFX72460.1 LJIJ01000222 ODN00228.1 SAUD01042376 RXG51071.1 CVRI01000055 CRL00964.1 KQ971338 EFA01759.1
AGBW02006209 OWR55239.1 PCG67856.1 KQ460817 KPJ12129.1 BABH01027215 KQ459337 KPJ01376.1 KQ982402 KYQ56858.1 ADTU01001056 ADTU01001057 ADTU01001058 ADTU01001059 ADTU01001060 ADTU01001061 GL437116 EFN71056.1 QOIP01000012 RLU15847.1 GL448228 EFN85056.1 NEVH01025635 PNF15379.1 KZ288192 PBC34298.1 JR038208 AEY58158.1 GEDC01013039 JAS24259.1 GEDC01022319 JAS14979.1 JXUM01031210 KQ560913 KXJ80367.1 GBYB01007492 GBYB01007494 JAG77259.1 JAG77261.1 GFDL01001212 JAV33833.1 ATLV01017581 KE525174 KFB42287.1 GEDC01019368 JAS17930.1 AXCM01002228 GDIQ01005978 JAN88759.1 GDIQ01117697 JAL34029.1 GDIP01228083 JAI95318.1 GDIP01070605 JAM33110.1 GDIP01228084 JAI95317.1 GECU01027695 JAS80011.1 GDIP01252332 LRGB01000872 JAI71069.1 KZS15433.1 GDIP01188990 JAJ34412.1 CH477401 GL888002 EGI69298.1 KQ414572 KOC71328.1 GECZ01007685 JAS62084.1 KQ435850 KOX70902.1 AJVK01010334 AJVK01010335 AXCN02000653 GGFK01003089 MBW36410.1 APCN01001690 AAAB01008980 GFDF01010486 JAV03598.1 ADMH02002138 ETN58343.1 GGFM01001964 MBW22715.1 GGFJ01003778 MBW52919.1 DS235243 EEB13855.1 GEZM01057056 JAV72474.1 GDIP01012526 JAM91189.1 KQ977720 KYN00496.1 GDAI01002062 JAI15541.1 GDIP01126305 JAL77409.1 KQ981855 KYN34855.1 GANO01000613 JAB59258.1 KQ980275 KYN17007.1 KK107488 EZA49910.1 UFQT01000023 SSX18021.1 ABLF02017887 ABLF02017889 ABLF02062618 GDIQ01227108 JAK24617.1 KQ434778 KZC04369.1 GDIP01144090 JAJ79312.1 GGLE01001541 MBY05667.1 GFJQ02004891 JAW02079.1 GDIQ01124086 JAL27640.1 GDRN01067921 JAI64295.1 GFPF01005553 MAA16699.1 GEDV01006170 JAP82387.1 KQ976418 KYM89496.1 GGMR01018042 MBY30661.1 ACPB03006490 PYGN01001092 PSN37674.1 GL732599 EFX72460.1 LJIJ01000222 ODN00228.1 SAUD01042376 RXG51071.1 CVRI01000055 CRL00964.1 KQ971338 EFA01759.1
Proteomes
UP000283053
UP000218220
UP000007151
UP000053240
UP000005204
UP000053268
+ More
UP000075809 UP000005205 UP000000311 UP000279307 UP000008237 UP000005203 UP000002358 UP000235965 UP000242457 UP000069940 UP000249989 UP000030765 UP000075880 UP000075883 UP000075900 UP000076858 UP000075920 UP000076408 UP000008820 UP000007755 UP000053825 UP000053105 UP000092462 UP000075886 UP000075884 UP000075881 UP000075840 UP000075885 UP000075882 UP000075903 UP000000673 UP000069272 UP000076407 UP000007062 UP000009046 UP000075902 UP000078542 UP000078541 UP000078492 UP000053097 UP000192223 UP000007819 UP000076502 UP000078540 UP000015103 UP000245037 UP000000305 UP000094527 UP000288706 UP000183832 UP000007266 UP000095300
UP000075809 UP000005205 UP000000311 UP000279307 UP000008237 UP000005203 UP000002358 UP000235965 UP000242457 UP000069940 UP000249989 UP000030765 UP000075880 UP000075883 UP000075900 UP000076858 UP000075920 UP000076408 UP000008820 UP000007755 UP000053825 UP000053105 UP000092462 UP000075886 UP000075884 UP000075881 UP000075840 UP000075885 UP000075882 UP000075903 UP000000673 UP000069272 UP000076407 UP000007062 UP000009046 UP000075902 UP000078542 UP000078541 UP000078492 UP000053097 UP000192223 UP000007819 UP000076502 UP000078540 UP000015103 UP000245037 UP000000305 UP000094527 UP000288706 UP000183832 UP000007266 UP000095300
Interpro
Gene 3D
ProteinModelPortal
A0A2H1WTS1
A0A3S2M6T9
A0A2A4J6Z2
A0A212FNF2
A0A2A4J7W9
A0A0N0PBT8
+ More
H9JIH3 A0A194QD48 A0A151X995 A0A158P1G7 E2A676 A0A3L8D5T8 E2BGS4 A0A088AUI8 K7J4H0 A0A2J7PGC3 A0A2A3ERD7 V9IAL3 A0A1B6DF01 A0A1B6CNC1 A0A182HAH3 A0A0C9QUR3 A0A1Q3G210 A0A084VWE3 A0A1B6CWT0 A0A182IWZ4 A0A182LYP8 A0A0P6JJ89 A0A0P5Q8I8 A0A0P4ZBF1 A0A182RRC7 A0A0P5XKV4 A0A1S4FEC3 A0A0P4YIZ8 A0A1B6HZ81 A0A0P4WQ01 A0A182W8V8 A0A0P5B673 A0A182Y6D8 Q175F8 F4W9B6 A0A0L7RKI4 A0A1B6GI18 A0A0N0U415 A0A1B0D1N2 A0A182QM25 A0A182NAW7 A0A182K4N2 A0A2M4A6P4 A0A182HRD4 A0A453YVI5 A0A182PNH1 A0A1S4H2H3 A0A182KZ01 A0A182VBT9 A0A1L8DAW2 W5J4J7 A0A2M3Z2I2 A0A2M4BIM9 A0A182F4C5 A0A182XHU2 Q7Q161 E0VKD9 A0A1Y1LFR7 A0A182TI87 A0A0P6BML2 A0A195CIC9 A0A0K8TN33 A0A0P5TIF6 A0A195F2Q6 U5EZK8 A0A151J3V6 A0A026W1G1 A0A1W4WU51 A0A336LMM2 J9JU51 A0A0P5HI90 A0A154NXI6 A0A0P5EAW3 A0A2R5L8C0 A0A1Z5L400 A0A0P5QGV7 A0A0P4WCV3 A0A224YLT0 A0A131YTV8 A0A195BRB1 A0A2S2PMH5 T1HKM5 A0A2P8Y0H3 E9H750 A0A1D2N4N3 A0A444SCH0 A0A1J1IN33 A0A1S4FWH1 D2A023 A0A1I8Q3G3 A0A1I8Q3G5 A0A1I8Q3E7
H9JIH3 A0A194QD48 A0A151X995 A0A158P1G7 E2A676 A0A3L8D5T8 E2BGS4 A0A088AUI8 K7J4H0 A0A2J7PGC3 A0A2A3ERD7 V9IAL3 A0A1B6DF01 A0A1B6CNC1 A0A182HAH3 A0A0C9QUR3 A0A1Q3G210 A0A084VWE3 A0A1B6CWT0 A0A182IWZ4 A0A182LYP8 A0A0P6JJ89 A0A0P5Q8I8 A0A0P4ZBF1 A0A182RRC7 A0A0P5XKV4 A0A1S4FEC3 A0A0P4YIZ8 A0A1B6HZ81 A0A0P4WQ01 A0A182W8V8 A0A0P5B673 A0A182Y6D8 Q175F8 F4W9B6 A0A0L7RKI4 A0A1B6GI18 A0A0N0U415 A0A1B0D1N2 A0A182QM25 A0A182NAW7 A0A182K4N2 A0A2M4A6P4 A0A182HRD4 A0A453YVI5 A0A182PNH1 A0A1S4H2H3 A0A182KZ01 A0A182VBT9 A0A1L8DAW2 W5J4J7 A0A2M3Z2I2 A0A2M4BIM9 A0A182F4C5 A0A182XHU2 Q7Q161 E0VKD9 A0A1Y1LFR7 A0A182TI87 A0A0P6BML2 A0A195CIC9 A0A0K8TN33 A0A0P5TIF6 A0A195F2Q6 U5EZK8 A0A151J3V6 A0A026W1G1 A0A1W4WU51 A0A336LMM2 J9JU51 A0A0P5HI90 A0A154NXI6 A0A0P5EAW3 A0A2R5L8C0 A0A1Z5L400 A0A0P5QGV7 A0A0P4WCV3 A0A224YLT0 A0A131YTV8 A0A195BRB1 A0A2S2PMH5 T1HKM5 A0A2P8Y0H3 E9H750 A0A1D2N4N3 A0A444SCH0 A0A1J1IN33 A0A1S4FWH1 D2A023 A0A1I8Q3G3 A0A1I8Q3G5 A0A1I8Q3E7
PDB
6IIT
E-value=1.45442e-09,
Score=149
Ontologies
GO
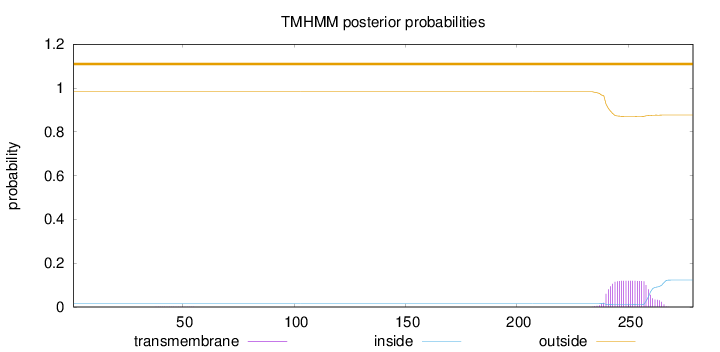
Topology
Length:
279
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.46077
Exp number, first 60 AAs:
0.00579
Total prob of N-in:
0.01695
outside
1 - 279
Population Genetic Test Statistics
Pi
232.99073
Theta
205.1
Tajima's D
0.574353
CLR
0.392569
CSRT
0.529573521323934
Interpretation
Uncertain
