Gene
KWMTBOMO08536
Pre Gene Modal
BGIBMGA009370
Annotation
PREDICTED:_uncharacterized_protein_ZC262.3_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.169
Sequence
CDS
ATGCGGTGGCTGCAGGCAGTATTGTCAGTAGCAGCTAGTTGCGCGGCTGTGGGGGTGGCGGTCTGTCCTGACACTTGCACGTGCGACGATGAACACCTGACCTGCTCCAATACGTCCGTCCAGGACAACAAGCTAACTAAGACGACGCTCGCCAAATACGGAAACCTCTTAGATACAATAATCTGGACAAACTCTAATATTAACTACCTCGAAATTGATTTGTTCAAAACATTTCCAAGACTGTCGTACATCGATTTGAGCGGAAACGTTATCAAGAAGACTGGAAACGGCCTGTTCGCGCAGTTCGGTCGACTGATTTACTTGAACATTTCCAGGAACGAACTAGAGGATATACCGAGGTCCACGTTCGCCGATTTAGAAAATCTAGAAGTCCTAGATCTGTCCCATAACCGGCTCAGATTCATACCGTTTCAAGTTTTCGAACCCATGCCCAAACTTCAGTACCTGGATTTATCGTATAATAAATTAGCAACATTTTTAGATTACTATTTCAATCCAAACAAACACTTAAAAACGCTGTTCCTAAACGATAATAGCCTAGTCAAAATAACATCTAACGCTCTAGTCAACTTGAAGGAGTTGGAAACATTGGATTTATCAAATAACAAATTAGACTACATACCGAAATCATTTTTCGACAATTTTGAATTGCTGAGGGAACTTAACTTAAGTTACAATCCTTTTCAAAACATATCACAAGACGCTTTCAAGAACCTCAAGAAACTGCTGTGGTTGAACATGAGCGGCAACAGACTACGGCTGCTTCCTCCGGTGCTCTTCCAGTATAACGAAAACCTTCAATCTCTCTACCTCGATCACACTGAACTAACAGTTATACAAAATACTAATTTCAAAAATTTAAATAATCTCAAGAGACTTTACATTAGGAATAATTCGTATTTACGTGAAATAGAAACGTACGTATTCCAAGATACTCTCAATTTAACGCATTTAAATATTAGTTCGAACGCATTAACGTTCCTACCGTTGTCTTTGAGCATGCTGGACCAGCTCCGGGAGTTGTGGATAGGAGGGAACCCGTGGGCTTGCGACTGCCGAATGGCGTGGTTCGTAGACTGGGCCGAACCGAGACTGAGCAGAGGAATCATAAAGTCTGAACTGAGCTGCGGCCAGTCTTACCCGGGCGAGATGTTGCCGATACTGCGCCACACGAACTGCAAGCCGCCTCGACTGATCTCAAGCAGTCCGCTGACGCTGTATCGACTGCAGTCGGACGCGCTATTAGAATGCGAATTCGTCGGTAATCCGACGCCTTCGATAACATGGATCACGCCAACCAGGACCGTGTTCCACTGGAACCCAGACCCGATCACGCCAGACATATTCCACAAGCACGGCGTCGCCCACGATCAGTATTATAACGCGATAGACAATTCGAAATCGAGAGTGAGAGTTCTTGAAGACGGTTCGCTATTTATCGGGAATATACACAGGGAAGACTGTGGAAGTTACTTGTGTGTCGGAACCAATCCTTCGGCGAACGTTTCCGCCGAAGTAGTCTTGAATATAGATCCTATGACCATGTACGAAATTAAAATATACAGTTTGATGTGGGGAGCGATATGCGCTGCCGGTTTCCTGGGACTCACGCTATTGGCCCAGGCGTTGCGATACATATTTTTCAGGTTCCGATTGATGGAGACGTGTTGCAGTTGTTGCGGGTGCGTACGCCGCGACGCGCCGCGCTCGCGACAAATCTACAGTATGTTGGACAACATAGAACAATATAAGAGACAACAACTAGAAAAATTAAGAGAAAACTATGCGGTTCAGGTACATCGAATAAAAGAAAATTGTACACAACAGATGGAATGGATACAGAACAGTTACTCGAGTCAAGCAAGTCACCTCCGCAACATTAGAGACATTGGAACGAATCATTTGACCGCCATGAGGGACCAGTATTACGATCAGGTAAAGCGCGTGAGAGAATACTCGACCAGTCAACTGAACTGGGTGCGAGAGAATTACGTATTCCAAAGAAACAAAATAAGGAAGTTCAGCGCGCATCAGATCCTCCGCATCCGCGAGTCTTACAAGTACCAGCAGCAAACCCTGAACAAAGTCCTCGAGAACCTGCCGAGCTTGTACTTCGAGAATTGCCGCAGCGGCTCGTGCGGTCGCAGCGACTCCATGGCGTTCGACCCCGACGTCGAGGTCATCAACATGTACCTCAAGACCAAGATAGAGAAGCTGTCCAATCTCGCCAGCCCCATAGCGGACGACGAGAGCAAGATATCGGTGTACTACACGCCGACGGAGCGGTCCGTCAACTCGCGGCGCAGCTCGCCCGTGCCGCTGCCCGAGGGTATCCACATCAACATGATTGAGTGCGAGCCCGAGTTGTGCTTGGAGCTGCCGTCCACGTCGCGGGGGGCCGGTGTGCGGGGGGGCGCGGTGCGGGCGGAGGCGGCGCGGGGGGGTGCGGCGCGGGCGGAGGCGGTGCGGGGGGGCGCGGCGCGGGCGGGGGCGGGCAAGCCGGCGAGCGAGCCGCTGCTGGGCGGGTGGCGCGGCGGGTCGCTGGGCGGCAGCGCGAGCTCGCCGGAGCTGCGTGCGGCGCGCGTGCTGCTGGCGGTGGAGCTGTGCGACGACACGCGCTTGTAG
Protein
MRWLQAVLSVAASCAAVGVAVCPDTCTCDDEHLTCSNTSVQDNKLTKTTLAKYGNLLDTIIWTNSNINYLEIDLFKTFPRLSYIDLSGNVIKKTGNGLFAQFGRLIYLNISRNELEDIPRSTFADLENLEVLDLSHNRLRFIPFQVFEPMPKLQYLDLSYNKLATFLDYYFNPNKHLKTLFLNDNSLVKITSNALVNLKELETLDLSNNKLDYIPKSFFDNFELLRELNLSYNPFQNISQDAFKNLKKLLWLNMSGNRLRLLPPVLFQYNENLQSLYLDHTELTVIQNTNFKNLNNLKRLYIRNNSYLREIETYVFQDTLNLTHLNISSNALTFLPLSLSMLDQLRELWIGGNPWACDCRMAWFVDWAEPRLSRGIIKSELSCGQSYPGEMLPILRHTNCKPPRLISSSPLTLYRLQSDALLECEFVGNPTPSITWITPTRTVFHWNPDPITPDIFHKHGVAHDQYYNAIDNSKSRVRVLEDGSLFIGNIHREDCGSYLCVGTNPSANVSAEVVLNIDPMTMYEIKIYSLMWGAICAAGFLGLTLLAQALRYIFFRFRLMETCCSCCGCVRRDAPRSRQIYSMLDNIEQYKRQQLEKLRENYAVQVHRIKENCTQQMEWIQNSYSSQASHLRNIRDIGTNHLTAMRDQYYDQVKRVREYSTSQLNWVRENYVFQRNKIRKFSAHQILRIRESYKYQQQTLNKVLENLPSLYFENCRSGSCGRSDSMAFDPDVEVINMYLKTKIEKLSNLASPIADDESKISVYYTPTERSVNSRRSSPVPLPEGIHINMIECEPELCLELPSTSRGAGVRGGAVRAEAARGGAARAEAVRGGAARAGAGKPASEPLLGGWRGGSLGGSASSPELRAARVLLAVELCDDTRL
Summary
Uniprot
H9JIM0
A0A2A4J855
A0A2H1WTZ4
A0A0L7LLM9
A0A0N0PC16
A0A194Q756
+ More
S4PAY0 A0A212FNH2 A0A0T6BE79 D2A022 A0A1W4X4V8 U4UW73 A0A1W4WU63 A0A1Y1MCI5 A0A2J7PGC0 A0A2P8Y083 N6U051 A0A0C9RCB0 A0A195CJ85 A0A151X952 A0A195BRD7 A0A158P1F6 A0A0L7RK52 A0A151J3H7 F4W9B7 A0A195F444 A0A0M8ZW97 A0A154NZD8 E2BGS5 E2A675 A0A088AV70 E9IKY3 A0A2A3ET29 A0A0J7NDL0 A0A2M3YZW1 W5JUN8 A0A2M4CI04 A0A2M4CI08 A0A2M4CJA7 A0A182P4B2 A0A182IBM1 A0A182X5Y9 A0A1S4GYQ7 A0A182F0T7 A0A2M3ZZU1 A0A2M3ZZM8 A0A2M3ZZP3 A0A182ITH6 A0A026W4E0 A0A336LIE8 A0A336JZD6 A0A084VFE5 A0A182GT13 Q7Q8I8 A0A1S4FBF5 A0A023EYA0 A0A182Q9Z1 E0VKD8 A0A182RK79 A0A182WFJ2 A0A2M3Z0K6 A0A2Y9D0R1 A0A2M4CY70 A0A182KE16 A0A2M4BCL7 A0A2M4BCQ8 Q178Y0 A0A182TQL4 A0A182KMF7 A0A1L8E2Y2 A0A1L8E2N6 A0A1L8E350 A0A1L8E3L2 A0A182NTD1 A0A182YQI2 A0A182G215 A0A1Q3FU65 A0A1Q3FUN2 A0A1Q3FU49 A0A1Q3FU76 A0A1Q3FU89 A0A1Q3FU85 A0A1Q3FUC1 A0A1Q3FU44 A0A1Q3FUL7 A0A1S4JSV8 A0A1Q3FAU0 A0A1Q3FDT2 A0A1Q3FEH4 A0A1Q3FDB4 A0A1Q3FE96 A0A1Q3FEK1 B4JA57 B0WUQ3 A0A0L0C120 T1PMU4 A0A0K8VMS4 A0A034V9N0 W8B2S1
S4PAY0 A0A212FNH2 A0A0T6BE79 D2A022 A0A1W4X4V8 U4UW73 A0A1W4WU63 A0A1Y1MCI5 A0A2J7PGC0 A0A2P8Y083 N6U051 A0A0C9RCB0 A0A195CJ85 A0A151X952 A0A195BRD7 A0A158P1F6 A0A0L7RK52 A0A151J3H7 F4W9B7 A0A195F444 A0A0M8ZW97 A0A154NZD8 E2BGS5 E2A675 A0A088AV70 E9IKY3 A0A2A3ET29 A0A0J7NDL0 A0A2M3YZW1 W5JUN8 A0A2M4CI04 A0A2M4CI08 A0A2M4CJA7 A0A182P4B2 A0A182IBM1 A0A182X5Y9 A0A1S4GYQ7 A0A182F0T7 A0A2M3ZZU1 A0A2M3ZZM8 A0A2M3ZZP3 A0A182ITH6 A0A026W4E0 A0A336LIE8 A0A336JZD6 A0A084VFE5 A0A182GT13 Q7Q8I8 A0A1S4FBF5 A0A023EYA0 A0A182Q9Z1 E0VKD8 A0A182RK79 A0A182WFJ2 A0A2M3Z0K6 A0A2Y9D0R1 A0A2M4CY70 A0A182KE16 A0A2M4BCL7 A0A2M4BCQ8 Q178Y0 A0A182TQL4 A0A182KMF7 A0A1L8E2Y2 A0A1L8E2N6 A0A1L8E350 A0A1L8E3L2 A0A182NTD1 A0A182YQI2 A0A182G215 A0A1Q3FU65 A0A1Q3FUN2 A0A1Q3FU49 A0A1Q3FU76 A0A1Q3FU89 A0A1Q3FU85 A0A1Q3FUC1 A0A1Q3FU44 A0A1Q3FUL7 A0A1S4JSV8 A0A1Q3FAU0 A0A1Q3FDT2 A0A1Q3FEH4 A0A1Q3FDB4 A0A1Q3FE96 A0A1Q3FEK1 B4JA57 B0WUQ3 A0A0L0C120 T1PMU4 A0A0K8VMS4 A0A034V9N0 W8B2S1
Pubmed
EMBL
BABH01027213
BABH01027214
NWSH01002610
PCG67858.1
ODYU01010995
SOQ56452.1
+ More
JTDY01000649 KOB76335.1 KQ460817 KPJ12130.1 KQ459337 KPJ01377.1 GAIX01004821 JAA87739.1 AGBW02006209 OWR55240.1 LJIG01001336 KRT85604.1 KQ971338 EFA02458.2 KB632390 ERL94485.1 GEZM01036401 JAV82718.1 NEVH01025635 PNF15378.1 PYGN01001092 PSN37673.1 APGK01054578 APGK01054579 APGK01054580 APGK01054581 APGK01054582 APGK01054583 APGK01054584 APGK01054585 KB741250 ENN71907.1 GBYB01004571 JAG74338.1 KQ977720 KYN00497.1 KQ982402 KYQ56859.1 KQ976418 KYM89494.1 ADTU01001058 KQ414572 KOC71327.1 KQ980275 KYN17006.1 GL888002 EGI69299.1 KQ981855 KYN34854.1 KQ435850 KOX70903.1 KQ434778 KZC04368.1 GL448228 EFN85057.1 GL437116 EFN71055.1 GL764026 EFZ18720.1 KZ288192 PBC34299.1 LBMM01006464 KMQ90595.1 GGFM01001053 MBW21804.1 ADMH02000444 ETN66464.1 GGFL01000789 MBW64967.1 GGFL01000798 MBW64976.1 GGFL01000790 MBW64968.1 APCN01000710 AAAB01008944 GGFK01000704 MBW34025.1 GGFK01000703 MBW34024.1 GGFK01000702 MBW34023.1 KK107488 QOIP01000012 EZA49909.1 RLU15842.1 UFQS01000017 UFQT01000017 SSW97478.1 SSX17864.1 SSW97477.1 SSX17863.1 ATLV01012384 KE524787 KFB36689.1 JXUM01085952 JXUM01085953 JXUM01085954 JXUM01085955 JXUM01085956 JXUM01085957 EAA10172.4 GAPW01000069 JAC13529.1 AXCN02000713 AAZO01003026 DS235243 EEB13854.1 GGFM01001260 MBW22011.1 GGFL01006105 MBW70283.1 GGFJ01001621 MBW50762.1 GGFJ01001620 MBW50761.1 CH477357 EAT42762.1 GFDF01001085 JAV12999.1 GFDF01001084 JAV13000.1 GFDF01000998 JAV13086.1 GFDF01000999 JAV13085.1 JXUM01006501 JXUM01006502 JXUM01006503 JXUM01006504 KQ560217 KXJ83769.1 GFDL01003905 JAV31140.1 GFDL01003902 JAV31143.1 GFDL01003921 JAV31124.1 GFDL01003895 JAV31150.1 GFDL01003920 JAV31125.1 GFDL01003918 JAV31127.1 GFDL01003907 JAV31138.1 GFDL01003904 JAV31141.1 GFDL01003903 JAV31142.1 GFDL01010427 JAV24618.1 GFDL01009324 JAV25721.1 GFDL01009085 JAV25960.1 GFDL01009498 JAV25547.1 GFDL01009171 JAV25874.1 GFDL01009093 JAV25952.1 CH916368 EDW03731.1 DS232108 EDS35039.1 JRES01001042 KNC26023.1 KA650092 AFP64721.1 GDHF01012135 JAI40179.1 GAKP01020706 GAKP01020704 JAC38248.1 GAMC01013713 JAB92842.1
JTDY01000649 KOB76335.1 KQ460817 KPJ12130.1 KQ459337 KPJ01377.1 GAIX01004821 JAA87739.1 AGBW02006209 OWR55240.1 LJIG01001336 KRT85604.1 KQ971338 EFA02458.2 KB632390 ERL94485.1 GEZM01036401 JAV82718.1 NEVH01025635 PNF15378.1 PYGN01001092 PSN37673.1 APGK01054578 APGK01054579 APGK01054580 APGK01054581 APGK01054582 APGK01054583 APGK01054584 APGK01054585 KB741250 ENN71907.1 GBYB01004571 JAG74338.1 KQ977720 KYN00497.1 KQ982402 KYQ56859.1 KQ976418 KYM89494.1 ADTU01001058 KQ414572 KOC71327.1 KQ980275 KYN17006.1 GL888002 EGI69299.1 KQ981855 KYN34854.1 KQ435850 KOX70903.1 KQ434778 KZC04368.1 GL448228 EFN85057.1 GL437116 EFN71055.1 GL764026 EFZ18720.1 KZ288192 PBC34299.1 LBMM01006464 KMQ90595.1 GGFM01001053 MBW21804.1 ADMH02000444 ETN66464.1 GGFL01000789 MBW64967.1 GGFL01000798 MBW64976.1 GGFL01000790 MBW64968.1 APCN01000710 AAAB01008944 GGFK01000704 MBW34025.1 GGFK01000703 MBW34024.1 GGFK01000702 MBW34023.1 KK107488 QOIP01000012 EZA49909.1 RLU15842.1 UFQS01000017 UFQT01000017 SSW97478.1 SSX17864.1 SSW97477.1 SSX17863.1 ATLV01012384 KE524787 KFB36689.1 JXUM01085952 JXUM01085953 JXUM01085954 JXUM01085955 JXUM01085956 JXUM01085957 EAA10172.4 GAPW01000069 JAC13529.1 AXCN02000713 AAZO01003026 DS235243 EEB13854.1 GGFM01001260 MBW22011.1 GGFL01006105 MBW70283.1 GGFJ01001621 MBW50762.1 GGFJ01001620 MBW50761.1 CH477357 EAT42762.1 GFDF01001085 JAV12999.1 GFDF01001084 JAV13000.1 GFDF01000998 JAV13086.1 GFDF01000999 JAV13085.1 JXUM01006501 JXUM01006502 JXUM01006503 JXUM01006504 KQ560217 KXJ83769.1 GFDL01003905 JAV31140.1 GFDL01003902 JAV31143.1 GFDL01003921 JAV31124.1 GFDL01003895 JAV31150.1 GFDL01003920 JAV31125.1 GFDL01003918 JAV31127.1 GFDL01003907 JAV31138.1 GFDL01003904 JAV31141.1 GFDL01003903 JAV31142.1 GFDL01010427 JAV24618.1 GFDL01009324 JAV25721.1 GFDL01009085 JAV25960.1 GFDL01009498 JAV25547.1 GFDL01009171 JAV25874.1 GFDL01009093 JAV25952.1 CH916368 EDW03731.1 DS232108 EDS35039.1 JRES01001042 KNC26023.1 KA650092 AFP64721.1 GDHF01012135 JAI40179.1 GAKP01020706 GAKP01020704 JAC38248.1 GAMC01013713 JAB92842.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053240
UP000053268
UP000007151
+ More
UP000007266 UP000192223 UP000030742 UP000235965 UP000245037 UP000019118 UP000078542 UP000075809 UP000078540 UP000005205 UP000053825 UP000078492 UP000007755 UP000078541 UP000053105 UP000076502 UP000008237 UP000000311 UP000005203 UP000242457 UP000036403 UP000000673 UP000075885 UP000075840 UP000076407 UP000069272 UP000075880 UP000053097 UP000279307 UP000030765 UP000069940 UP000007062 UP000075886 UP000009046 UP000075900 UP000075920 UP000075881 UP000008820 UP000075902 UP000075882 UP000075884 UP000076408 UP000249989 UP000001070 UP000002320 UP000037069
UP000007266 UP000192223 UP000030742 UP000235965 UP000245037 UP000019118 UP000078542 UP000075809 UP000078540 UP000005205 UP000053825 UP000078492 UP000007755 UP000078541 UP000053105 UP000076502 UP000008237 UP000000311 UP000005203 UP000242457 UP000036403 UP000000673 UP000075885 UP000075840 UP000076407 UP000069272 UP000075880 UP000053097 UP000279307 UP000030765 UP000069940 UP000007062 UP000075886 UP000009046 UP000075900 UP000075920 UP000075881 UP000008820 UP000075902 UP000075882 UP000075884 UP000076408 UP000249989 UP000001070 UP000002320 UP000037069
PRIDE
Interpro
SUPFAM
SSF48726
SSF48726
Gene 3D
ProteinModelPortal
H9JIM0
A0A2A4J855
A0A2H1WTZ4
A0A0L7LLM9
A0A0N0PC16
A0A194Q756
+ More
S4PAY0 A0A212FNH2 A0A0T6BE79 D2A022 A0A1W4X4V8 U4UW73 A0A1W4WU63 A0A1Y1MCI5 A0A2J7PGC0 A0A2P8Y083 N6U051 A0A0C9RCB0 A0A195CJ85 A0A151X952 A0A195BRD7 A0A158P1F6 A0A0L7RK52 A0A151J3H7 F4W9B7 A0A195F444 A0A0M8ZW97 A0A154NZD8 E2BGS5 E2A675 A0A088AV70 E9IKY3 A0A2A3ET29 A0A0J7NDL0 A0A2M3YZW1 W5JUN8 A0A2M4CI04 A0A2M4CI08 A0A2M4CJA7 A0A182P4B2 A0A182IBM1 A0A182X5Y9 A0A1S4GYQ7 A0A182F0T7 A0A2M3ZZU1 A0A2M3ZZM8 A0A2M3ZZP3 A0A182ITH6 A0A026W4E0 A0A336LIE8 A0A336JZD6 A0A084VFE5 A0A182GT13 Q7Q8I8 A0A1S4FBF5 A0A023EYA0 A0A182Q9Z1 E0VKD8 A0A182RK79 A0A182WFJ2 A0A2M3Z0K6 A0A2Y9D0R1 A0A2M4CY70 A0A182KE16 A0A2M4BCL7 A0A2M4BCQ8 Q178Y0 A0A182TQL4 A0A182KMF7 A0A1L8E2Y2 A0A1L8E2N6 A0A1L8E350 A0A1L8E3L2 A0A182NTD1 A0A182YQI2 A0A182G215 A0A1Q3FU65 A0A1Q3FUN2 A0A1Q3FU49 A0A1Q3FU76 A0A1Q3FU89 A0A1Q3FU85 A0A1Q3FUC1 A0A1Q3FU44 A0A1Q3FUL7 A0A1S4JSV8 A0A1Q3FAU0 A0A1Q3FDT2 A0A1Q3FEH4 A0A1Q3FDB4 A0A1Q3FE96 A0A1Q3FEK1 B4JA57 B0WUQ3 A0A0L0C120 T1PMU4 A0A0K8VMS4 A0A034V9N0 W8B2S1
S4PAY0 A0A212FNH2 A0A0T6BE79 D2A022 A0A1W4X4V8 U4UW73 A0A1W4WU63 A0A1Y1MCI5 A0A2J7PGC0 A0A2P8Y083 N6U051 A0A0C9RCB0 A0A195CJ85 A0A151X952 A0A195BRD7 A0A158P1F6 A0A0L7RK52 A0A151J3H7 F4W9B7 A0A195F444 A0A0M8ZW97 A0A154NZD8 E2BGS5 E2A675 A0A088AV70 E9IKY3 A0A2A3ET29 A0A0J7NDL0 A0A2M3YZW1 W5JUN8 A0A2M4CI04 A0A2M4CI08 A0A2M4CJA7 A0A182P4B2 A0A182IBM1 A0A182X5Y9 A0A1S4GYQ7 A0A182F0T7 A0A2M3ZZU1 A0A2M3ZZM8 A0A2M3ZZP3 A0A182ITH6 A0A026W4E0 A0A336LIE8 A0A336JZD6 A0A084VFE5 A0A182GT13 Q7Q8I8 A0A1S4FBF5 A0A023EYA0 A0A182Q9Z1 E0VKD8 A0A182RK79 A0A182WFJ2 A0A2M3Z0K6 A0A2Y9D0R1 A0A2M4CY70 A0A182KE16 A0A2M4BCL7 A0A2M4BCQ8 Q178Y0 A0A182TQL4 A0A182KMF7 A0A1L8E2Y2 A0A1L8E2N6 A0A1L8E350 A0A1L8E3L2 A0A182NTD1 A0A182YQI2 A0A182G215 A0A1Q3FU65 A0A1Q3FUN2 A0A1Q3FU49 A0A1Q3FU76 A0A1Q3FU89 A0A1Q3FU85 A0A1Q3FUC1 A0A1Q3FU44 A0A1Q3FUL7 A0A1S4JSV8 A0A1Q3FAU0 A0A1Q3FDT2 A0A1Q3FEH4 A0A1Q3FDB4 A0A1Q3FE96 A0A1Q3FEK1 B4JA57 B0WUQ3 A0A0L0C120 T1PMU4 A0A0K8VMS4 A0A034V9N0 W8B2S1
PDB
3ZYO
E-value=4.64984e-32,
Score=348
Ontologies
GO
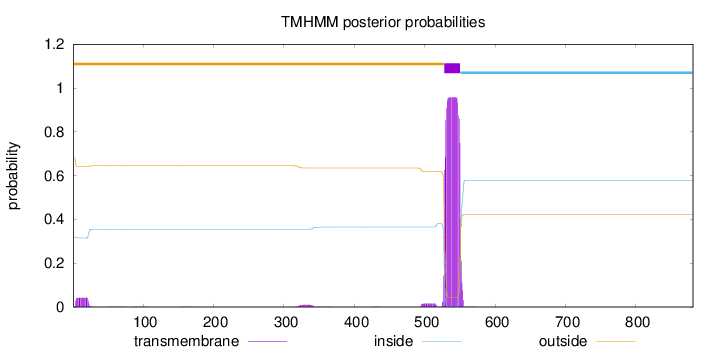
Topology
SignalP
Position: 1 - 16,
Likelihood: 0.997229
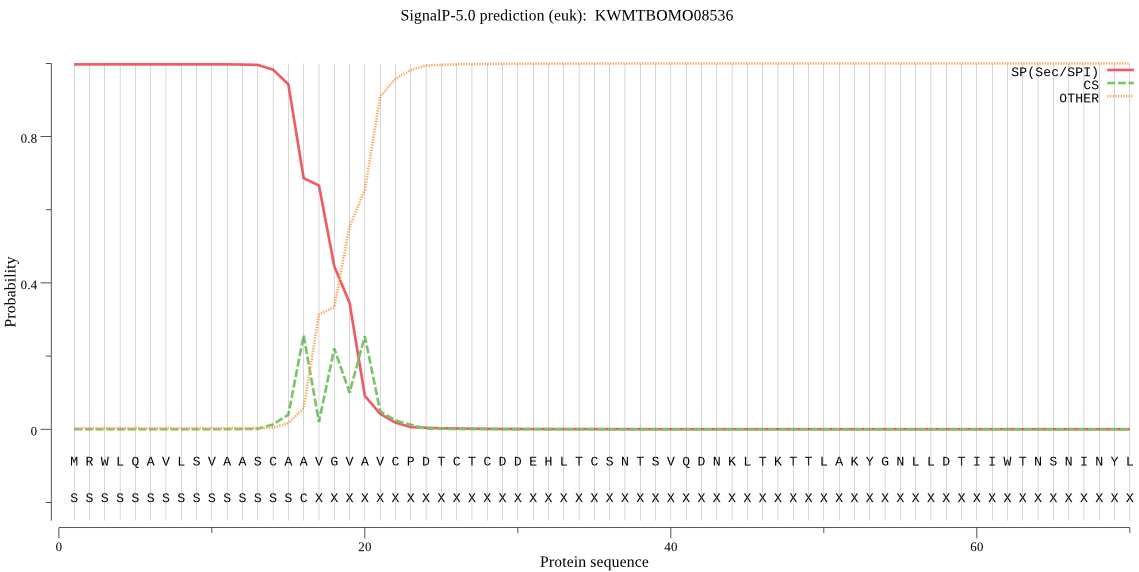
Length:
881
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.05409
Exp number, first 60 AAs:
0.80391
Total prob of N-in:
0.31803
outside
1 - 527
TMhelix
528 - 550
inside
551 - 881
Population Genetic Test Statistics
Pi
170.907146
Theta
168.868854
Tajima's D
0.010102
CLR
10.400055
CSRT
0.373031348432578
Interpretation
Uncertain
