Gene
KWMTBOMO08535 Validated by peptides from experiments
Annotation
PREDICTED:_putative_oxidoreductase_GLYR1_homolog_isoform_X3_[Papilio_xuthus]
Full name
Putative oxidoreductase GLYR1 homolog
Alternative Name
Glyoxylate reductase 1 homolog
Nuclear protein NP60 homolog
Nuclear protein NP60 homolog
Location in the cell
Cytoplasmic Reliability : 2.305
Sequence
CDS
ATGTGCAAAGACTTCGAGAAAGTTGGGGCCACGATTGCGGTGACACCATGCGATGTGGTGGAAGAAGCCGACATCACGTTCTCATGTGTGGCTGACCCTCAAGCAGCTAAAGAGATGGTGTTCGGTAATTGTGGCGTGCTGCACTGCCCGACGCTGGAGTCCAAGGGCTACGTCGAGATGACCTCGATCGACGCCGACACCTCGCACGACATCGTGGAAGCCATCAGCGGCAAGGGTGGCCGCTACCTGGAGGCCCAGATACAAGGATCGAAAACGCAAGCGGAGGAAGGCACGCTGATAGTGCTGGCGGCCGGCGACCGCTCGCTGTTCGACGACTGCCAGTCCTGCTTCAAGGCGATGAGCAAGAACTCGTTTTACCTTGGTAGTGAGATCGGCAACGCGTCGAAGATGAACTCCGTGCTGCAGGTGGTGGGCGGCGTGTCGCTGGCCGCGCTGGCCGAGGGGCTGGCGCTGGCCGACCGCGCGGGGCTCAGCCAGGCCGACCTGCTCGACGTGCTGGCCCTCACGCCGCTCGCCTCCCCGCACCTCATCCTCAAAGGACGCGCCATGATCGAGTCCTCCTACTCCACGCACCAGCCCCTCACGCACATGCAGAAGGACCTTAAGCTGGCGCTGGGGCTGGGCGACGCGCTGGAGCAGTCCCTGCCGCTCACGGCCACCACCAACGAGATCTTCAAGCACGCCAAGCGCCTCGGCTACGCGAACCACGACGTGGCCGCCGTCTACATCCGCGCCCGCTTCTGA
Protein
MCKDFEKVGATIAVTPCDVVEEADITFSCVADPQAAKEMVFGNCGVLHCPTLESKGYVEMTSIDADTSHDIVEAISGKGGRYLEAQIQGSKTQAEEGTLIVLAAGDRSLFDDCQSCFKAMSKNSFYLGSEIGNASKMNSVLQVVGGVSLAALAEGLALADRAGLSQADLLDVLALTPLASPHLILKGRAMIESSYSTHQPLTHMQKDLKLALGLGDALEQSLPLTATTNEIFKHAKRLGYANHDVAAVYIRARF
Summary
Description
May have oxidoreductase activity.
Miscellaneous
The conserved NAD-binding sites suggest that this protein may have oxidoreductase activity.
Similarity
Belongs to the HIBADH-related family. NP60 subfamily.
Keywords
Complete proteome
NAD
Oxidoreductase
Reference proteome
Feature
chain Putative oxidoreductase GLYR1 homolog
Uniprot
A0A212FPF0
A0A2A4JAC5
A0A2H1VKG6
A0A3S2M6T9
A0A0L7KJ14
A0A0N1IDA5
+ More
A0A194Q8T3 A0A067QXW9 A0A1B6MMY6 A0A1B6KHL1 E9IKY4 A0A0L7RKQ1 A0A154NXM1 E2BGS6 A0A026W1K6 A0A1B6GI18 A0A088AUI9 A0A0J7KTK1 E9H752 E0VKD7 A0A1B6FJJ2 A0A195BT00 A0A195CIF2 A0A1B6CWT0 A0A195F314 A0A1S4ESI7 A0A232EL63 A0A423SFH2 A0A151J3H2 A0A0P5EUM9 A0A2A3ERD7 V9IAL3 A0A3L8D5T8 A0A0P4WCV3 A0A0P4W762 A0A0C9QUR3 A0A336LMM2 A0A158P1G7 A0A0P5HI90 A0A0P5QGV7 A0A0P5EAW3 A0A0P4WQ01 A0A0P6JJ89 A0A0P5TIF6 A0A182SZT5 A0A0P6BML2 K7J4H0 T1JPD6 A0A0K2TU36 A0A0N0BDT7 A0A1L8DAW2 A0A1B0D1N2 F4W9B8 A0A1B0CTI4 A0A182NAW7 A0A182W8V8 A0A084VWE3 A0A182HRD4 A0A182RRC7 A0A182XHU2 A0A182K4N2 A0A182QM25 A0A182VBT9 A0A0P5B673 A0A182TI87 A0A182LYP8 A0A182Y6D8 A0A0K8TN33 A0A453YVI5 A0A1Q3G210 A0A182PNH1 A0A1S4H2H3 A0A182KZ01 A0A182IWZ4 Q7Q161 A0A182HAH3 A0A443REM0 B0W036 A0A443S6D6 U5EZK8 A0A1S4FEC3 Q175F8 T1EAE6 A0A2M4A6P4 W5J4J7 A0A2M3Z2I2 A0A2P8Y0A6 A0A2J7PG37 A0A2M4BIM9 A0A1B6KTN4 A0A444SCD3 Q16LQ1 A0A293M635 A0A182F4C5 T1KD49 Q175A9 V5HVK5 A0A131Y2N9 A0A2R5L8C0 A0A1S4FEK4
A0A194Q8T3 A0A067QXW9 A0A1B6MMY6 A0A1B6KHL1 E9IKY4 A0A0L7RKQ1 A0A154NXM1 E2BGS6 A0A026W1K6 A0A1B6GI18 A0A088AUI9 A0A0J7KTK1 E9H752 E0VKD7 A0A1B6FJJ2 A0A195BT00 A0A195CIF2 A0A1B6CWT0 A0A195F314 A0A1S4ESI7 A0A232EL63 A0A423SFH2 A0A151J3H2 A0A0P5EUM9 A0A2A3ERD7 V9IAL3 A0A3L8D5T8 A0A0P4WCV3 A0A0P4W762 A0A0C9QUR3 A0A336LMM2 A0A158P1G7 A0A0P5HI90 A0A0P5QGV7 A0A0P5EAW3 A0A0P4WQ01 A0A0P6JJ89 A0A0P5TIF6 A0A182SZT5 A0A0P6BML2 K7J4H0 T1JPD6 A0A0K2TU36 A0A0N0BDT7 A0A1L8DAW2 A0A1B0D1N2 F4W9B8 A0A1B0CTI4 A0A182NAW7 A0A182W8V8 A0A084VWE3 A0A182HRD4 A0A182RRC7 A0A182XHU2 A0A182K4N2 A0A182QM25 A0A182VBT9 A0A0P5B673 A0A182TI87 A0A182LYP8 A0A182Y6D8 A0A0K8TN33 A0A453YVI5 A0A1Q3G210 A0A182PNH1 A0A1S4H2H3 A0A182KZ01 A0A182IWZ4 Q7Q161 A0A182HAH3 A0A443REM0 B0W036 A0A443S6D6 U5EZK8 A0A1S4FEC3 Q175F8 T1EAE6 A0A2M4A6P4 W5J4J7 A0A2M3Z2I2 A0A2P8Y0A6 A0A2J7PG37 A0A2M4BIM9 A0A1B6KTN4 A0A444SCD3 Q16LQ1 A0A293M635 A0A182F4C5 T1KD49 Q175A9 V5HVK5 A0A131Y2N9 A0A2R5L8C0 A0A1S4FEK4
EC Number
1.-.-.-
Pubmed
EMBL
AGBW02002847
OWR55607.1
NWSH01002408
PCG68353.1
ODYU01003037
SOQ41313.1
+ More
RSAL01000023 RVE52294.1 JTDY01009677 KOB63066.1 KQ459243 KPJ02449.1 KQ459337 KPJ01380.1 KK853227 KDR09657.1 GEBQ01002748 JAT37229.1 GEBQ01029020 JAT10957.1 GL764026 EFZ18698.1 KQ414572 KOC71326.1 KQ434778 KZC04367.1 GL448228 EFN85058.1 KK107488 EZA49908.1 GECZ01007685 JAS62084.1 LBMM01003337 KMQ93646.1 GL732599 EFX72461.1 DS235243 EEB13853.1 GECZ01019403 JAS50366.1 KQ976418 KYM89493.1 KQ977720 KYN00498.1 GEDC01019368 JAS17930.1 KQ981855 KYN34853.1 NNAY01003627 OXU19085.1 QCYY01003519 ROT62952.1 KQ980275 KYN17005.1 GDIQ01267025 JAJ84699.1 KZ288192 PBC34298.1 JR038208 AEY58158.1 QOIP01000012 RLU15847.1 GDRN01067921 JAI64295.1 GDRN01067922 JAI64294.1 GBYB01007492 GBYB01007494 JAG77259.1 JAG77261.1 UFQT01000023 SSX18021.1 ADTU01001056 ADTU01001057 ADTU01001058 ADTU01001059 ADTU01001060 ADTU01001061 GDIQ01227108 JAK24617.1 GDIQ01124086 JAL27640.1 GDIP01144090 JAJ79312.1 GDIP01252332 LRGB01000872 JAI71069.1 KZS15433.1 GDIQ01005978 JAN88759.1 GDIP01126305 JAL77409.1 GDIP01012526 JAM91189.1 JH431826 HACA01011540 CDW28901.1 KQ435850 KOX70904.1 GFDF01010486 JAV03598.1 AJVK01010334 AJVK01010335 GL888002 EGI69300.1 AJWK01027661 ATLV01017581 KE525174 KFB42287.1 APCN01001690 AXCN02000653 GDIP01188990 JAJ34412.1 AXCM01002228 GDAI01002062 JAI15541.1 AAAB01008980 GFDL01001212 JAV33833.1 JXUM01031210 KQ560913 KXJ80367.1 NCKU01000903 RWS13714.1 DS231816 EDS37608.1 NCKV01007112 RWS23106.1 GANO01000613 JAB59258.1 CH477401 GAMD01000497 JAB01094.1 GGFK01003089 MBW36410.1 ADMH02002138 ETN58343.1 GGFM01001964 MBW22715.1 PYGN01001092 PSN37676.1 NEVH01025635 PNF15293.1 GGFJ01003778 MBW52919.1 GEBQ01025161 JAT14816.1 SAUD01042376 RXG51069.1 CH477898 EAT35237.1 GFWV01011495 MAA36224.1 CAEY01002009 CH477404 EAT41622.1 GANP01004181 JAB80287.1 GEFM01003051 JAP72745.1 GGLE01001541 MBY05667.1
RSAL01000023 RVE52294.1 JTDY01009677 KOB63066.1 KQ459243 KPJ02449.1 KQ459337 KPJ01380.1 KK853227 KDR09657.1 GEBQ01002748 JAT37229.1 GEBQ01029020 JAT10957.1 GL764026 EFZ18698.1 KQ414572 KOC71326.1 KQ434778 KZC04367.1 GL448228 EFN85058.1 KK107488 EZA49908.1 GECZ01007685 JAS62084.1 LBMM01003337 KMQ93646.1 GL732599 EFX72461.1 DS235243 EEB13853.1 GECZ01019403 JAS50366.1 KQ976418 KYM89493.1 KQ977720 KYN00498.1 GEDC01019368 JAS17930.1 KQ981855 KYN34853.1 NNAY01003627 OXU19085.1 QCYY01003519 ROT62952.1 KQ980275 KYN17005.1 GDIQ01267025 JAJ84699.1 KZ288192 PBC34298.1 JR038208 AEY58158.1 QOIP01000012 RLU15847.1 GDRN01067921 JAI64295.1 GDRN01067922 JAI64294.1 GBYB01007492 GBYB01007494 JAG77259.1 JAG77261.1 UFQT01000023 SSX18021.1 ADTU01001056 ADTU01001057 ADTU01001058 ADTU01001059 ADTU01001060 ADTU01001061 GDIQ01227108 JAK24617.1 GDIQ01124086 JAL27640.1 GDIP01144090 JAJ79312.1 GDIP01252332 LRGB01000872 JAI71069.1 KZS15433.1 GDIQ01005978 JAN88759.1 GDIP01126305 JAL77409.1 GDIP01012526 JAM91189.1 JH431826 HACA01011540 CDW28901.1 KQ435850 KOX70904.1 GFDF01010486 JAV03598.1 AJVK01010334 AJVK01010335 GL888002 EGI69300.1 AJWK01027661 ATLV01017581 KE525174 KFB42287.1 APCN01001690 AXCN02000653 GDIP01188990 JAJ34412.1 AXCM01002228 GDAI01002062 JAI15541.1 AAAB01008980 GFDL01001212 JAV33833.1 JXUM01031210 KQ560913 KXJ80367.1 NCKU01000903 RWS13714.1 DS231816 EDS37608.1 NCKV01007112 RWS23106.1 GANO01000613 JAB59258.1 CH477401 GAMD01000497 JAB01094.1 GGFK01003089 MBW36410.1 ADMH02002138 ETN58343.1 GGFM01001964 MBW22715.1 PYGN01001092 PSN37676.1 NEVH01025635 PNF15293.1 GGFJ01003778 MBW52919.1 GEBQ01025161 JAT14816.1 SAUD01042376 RXG51069.1 CH477898 EAT35237.1 GFWV01011495 MAA36224.1 CAEY01002009 CH477404 EAT41622.1 GANP01004181 JAB80287.1 GEFM01003051 JAP72745.1 GGLE01001541 MBY05667.1
Proteomes
UP000007151
UP000218220
UP000283053
UP000037510
UP000053268
UP000027135
+ More
UP000053825 UP000076502 UP000008237 UP000053097 UP000005203 UP000036403 UP000000305 UP000009046 UP000078540 UP000078542 UP000078541 UP000079169 UP000215335 UP000283509 UP000078492 UP000242457 UP000279307 UP000005205 UP000076858 UP000075901 UP000002358 UP000053105 UP000092462 UP000007755 UP000092461 UP000075884 UP000075920 UP000030765 UP000075840 UP000075900 UP000076407 UP000075881 UP000075886 UP000075903 UP000075902 UP000075883 UP000076408 UP000075885 UP000075882 UP000075880 UP000007062 UP000069940 UP000249989 UP000285301 UP000002320 UP000288716 UP000008820 UP000000673 UP000245037 UP000235965 UP000288706 UP000069272 UP000015104
UP000053825 UP000076502 UP000008237 UP000053097 UP000005203 UP000036403 UP000000305 UP000009046 UP000078540 UP000078542 UP000078541 UP000079169 UP000215335 UP000283509 UP000078492 UP000242457 UP000279307 UP000005205 UP000076858 UP000075901 UP000002358 UP000053105 UP000092462 UP000007755 UP000092461 UP000075884 UP000075920 UP000030765 UP000075840 UP000075900 UP000076407 UP000075881 UP000075886 UP000075903 UP000075902 UP000075883 UP000076408 UP000075885 UP000075882 UP000075880 UP000007062 UP000069940 UP000249989 UP000285301 UP000002320 UP000288716 UP000008820 UP000000673 UP000245037 UP000235965 UP000288706 UP000069272 UP000015104
Interpro
Gene 3D
ProteinModelPortal
A0A212FPF0
A0A2A4JAC5
A0A2H1VKG6
A0A3S2M6T9
A0A0L7KJ14
A0A0N1IDA5
+ More
A0A194Q8T3 A0A067QXW9 A0A1B6MMY6 A0A1B6KHL1 E9IKY4 A0A0L7RKQ1 A0A154NXM1 E2BGS6 A0A026W1K6 A0A1B6GI18 A0A088AUI9 A0A0J7KTK1 E9H752 E0VKD7 A0A1B6FJJ2 A0A195BT00 A0A195CIF2 A0A1B6CWT0 A0A195F314 A0A1S4ESI7 A0A232EL63 A0A423SFH2 A0A151J3H2 A0A0P5EUM9 A0A2A3ERD7 V9IAL3 A0A3L8D5T8 A0A0P4WCV3 A0A0P4W762 A0A0C9QUR3 A0A336LMM2 A0A158P1G7 A0A0P5HI90 A0A0P5QGV7 A0A0P5EAW3 A0A0P4WQ01 A0A0P6JJ89 A0A0P5TIF6 A0A182SZT5 A0A0P6BML2 K7J4H0 T1JPD6 A0A0K2TU36 A0A0N0BDT7 A0A1L8DAW2 A0A1B0D1N2 F4W9B8 A0A1B0CTI4 A0A182NAW7 A0A182W8V8 A0A084VWE3 A0A182HRD4 A0A182RRC7 A0A182XHU2 A0A182K4N2 A0A182QM25 A0A182VBT9 A0A0P5B673 A0A182TI87 A0A182LYP8 A0A182Y6D8 A0A0K8TN33 A0A453YVI5 A0A1Q3G210 A0A182PNH1 A0A1S4H2H3 A0A182KZ01 A0A182IWZ4 Q7Q161 A0A182HAH3 A0A443REM0 B0W036 A0A443S6D6 U5EZK8 A0A1S4FEC3 Q175F8 T1EAE6 A0A2M4A6P4 W5J4J7 A0A2M3Z2I2 A0A2P8Y0A6 A0A2J7PG37 A0A2M4BIM9 A0A1B6KTN4 A0A444SCD3 Q16LQ1 A0A293M635 A0A182F4C5 T1KD49 Q175A9 V5HVK5 A0A131Y2N9 A0A2R5L8C0 A0A1S4FEK4
A0A194Q8T3 A0A067QXW9 A0A1B6MMY6 A0A1B6KHL1 E9IKY4 A0A0L7RKQ1 A0A154NXM1 E2BGS6 A0A026W1K6 A0A1B6GI18 A0A088AUI9 A0A0J7KTK1 E9H752 E0VKD7 A0A1B6FJJ2 A0A195BT00 A0A195CIF2 A0A1B6CWT0 A0A195F314 A0A1S4ESI7 A0A232EL63 A0A423SFH2 A0A151J3H2 A0A0P5EUM9 A0A2A3ERD7 V9IAL3 A0A3L8D5T8 A0A0P4WCV3 A0A0P4W762 A0A0C9QUR3 A0A336LMM2 A0A158P1G7 A0A0P5HI90 A0A0P5QGV7 A0A0P5EAW3 A0A0P4WQ01 A0A0P6JJ89 A0A0P5TIF6 A0A182SZT5 A0A0P6BML2 K7J4H0 T1JPD6 A0A0K2TU36 A0A0N0BDT7 A0A1L8DAW2 A0A1B0D1N2 F4W9B8 A0A1B0CTI4 A0A182NAW7 A0A182W8V8 A0A084VWE3 A0A182HRD4 A0A182RRC7 A0A182XHU2 A0A182K4N2 A0A182QM25 A0A182VBT9 A0A0P5B673 A0A182TI87 A0A182LYP8 A0A182Y6D8 A0A0K8TN33 A0A453YVI5 A0A1Q3G210 A0A182PNH1 A0A1S4H2H3 A0A182KZ01 A0A182IWZ4 Q7Q161 A0A182HAH3 A0A443REM0 B0W036 A0A443S6D6 U5EZK8 A0A1S4FEC3 Q175F8 T1EAE6 A0A2M4A6P4 W5J4J7 A0A2M3Z2I2 A0A2P8Y0A6 A0A2J7PG37 A0A2M4BIM9 A0A1B6KTN4 A0A444SCD3 Q16LQ1 A0A293M635 A0A182F4C5 T1KD49 Q175A9 V5HVK5 A0A131Y2N9 A0A2R5L8C0 A0A1S4FEK4
PDB
2UYY
E-value=2.25774e-41,
Score=422
Ontologies
GO
Topology
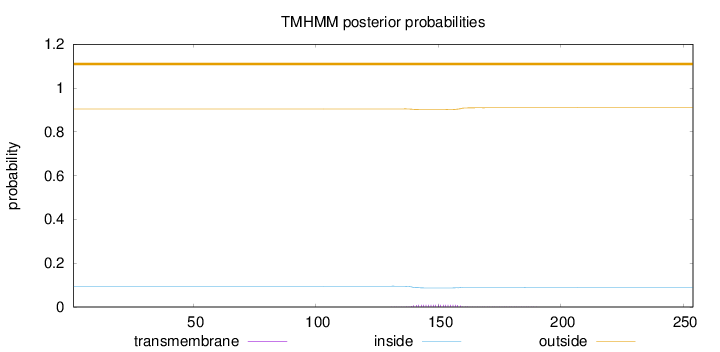
Length:
254
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.23082
Exp number, first 60 AAs:
0
Total prob of N-in:
0.09522
outside
1 - 254
Population Genetic Test Statistics
Pi
199.585372
Theta
147.352226
Tajima's D
1.076289
CLR
0.522448
CSRT
0.679366031698415
Interpretation
Uncertain
