Gene
KWMTBOMO08529
Pre Gene Modal
BGIBMGA009368
Annotation
PREDICTED:_uncharacterized_protein_LOC106713881_isoform_X1_[Papilio_machaon]
Location in the cell
Mitochondrial Reliability : 1.002 Nuclear Reliability : 1.406
Sequence
CDS
ATGGGTTGTTGCTATAGCATTTGCCACAAAGACAACGACCCACAGGAGAACGTTATAAACGAAAGGACACATCTACTAGAAGTAACGGAGCAGCAACAGGAGACGCTGATACCTGTGGTATCAGCGTCCACACCACCGAGGAAACCAGACGAACAGACTGCTTTGAATAGGATACTTCATGAGACTGCAAATAATGTGATCGACGTGGGCGCGCTGGGCCCGTACACACTGGAGGCCAGCGCGTACAGCGAGAGGGTGCGCGTGTACACGGCGCGGGTGGCGGCCGTGAGCGTGGCGGCCCCCCGCCCCGCGCCGCGGCTCCTCGCTGACGTCACCCACGCCGAGCGAGTGGCCCTACTCAGCTGTCCCCCAATACCGGCCGCAGATAAGGAGCTGATCAGTGGCGCAGTGAGACGAGCCGCCGCCGCCATATCGGAGCTCAGGGTCGAGCACCGGGAGGACCTGGTGGTGCCCTTCCGGGTCCCCTAA
Protein
MGCCYSICHKDNDPQENVINERTHLLEVTEQQQETLIPVVSASTPPRKPDEQTALNRILHETANNVIDVGALGPYTLEASAYSERVRVYTARVAAVSVAAPRPAPRLLADVTHAERVALLSCPPIPAADKELISGAVRRAAAAISELRVEHREDLVVPFRVP
Summary
Similarity
Belongs to the histone H1/H5 family.
Uniprot
A0A3S2NYL2
A0A0N1IF32
S4NRU2
A0A212FPH1
A0A194Q8U1
A0A2H1WIV9
+ More
A0A232EUK8 E2ADU9 K7J5X7 A0A0L7QJZ3 A0A151X682 A0A195E513 F4WH77 A0A151IH42 V9IGE6 A0A158NXG2 A0A3L8DXS3 A0A088AGW9 A0A195F3T8 A0A026WPE8 A0A0C9RZ94 A0A0M9A1H3 A0A0J7NA31 A0A2J7R3A9 A0A2A3ESC0 A0A195B9D0 D2A1I8 A0A067R1V2 A0A1Y1LZ56 A0A1B6DAU7 A0A1B6HBE3 V5GRP0 A0A154P6I7 A0A1B6EV42 A0A2M3ZDF1 W5JWC4 A0A2M4AYR2 A0A0A9WRN2 A0A0P4WF90 U5EWX5 A0A182FKA7 A0A2R7WZ06 A0A182YIR9 A0A2M4C115 A0A182STF4 A0A0P4VKV8 R4G8C9 A0A0V0G5I6 A0A182RJR3 A0A182JAE4 A0A2C9GRG2 A0A224XYC6 A0A182LNC5 Q7QDW9 A0A182Q4M6 A0A210QD27 A0A182W3V4 A0A023EJ00 Q17NY6 A7RFI0 R7V1T1 E0V989 A0A1W4WS63 A0A1B0CSX4 A0A1B6KAL9 A0A0K8TRA4 A0A0A9W3R7 A0A0P5GV55 A0A0P5KDY1 A0A1L8DV71 A0A2P8XVA5 E9G1G6 A0A023F7K8 V4A3J1 A0A182PSB2 A0A182NDN8 A0A336LIF4 A0A182X842 A0A182UCU3 A0A1Q3F8H1 A0A1Q3F8N7 C3YZ87 B0XCW5 A0A1J1HIU0 A0A0N7ZJC9 A0A0L8HLK3 A0A1S3D9S7 T1JL32 B3M3J9 A0A0L8HMM1 B3N2B1 A0A1D2NG19 A0A2R5LIG3 A0A0P6F091 G3VLI0 A0A293MV49 A0A2U3Z9V8 U4UIG1
A0A232EUK8 E2ADU9 K7J5X7 A0A0L7QJZ3 A0A151X682 A0A195E513 F4WH77 A0A151IH42 V9IGE6 A0A158NXG2 A0A3L8DXS3 A0A088AGW9 A0A195F3T8 A0A026WPE8 A0A0C9RZ94 A0A0M9A1H3 A0A0J7NA31 A0A2J7R3A9 A0A2A3ESC0 A0A195B9D0 D2A1I8 A0A067R1V2 A0A1Y1LZ56 A0A1B6DAU7 A0A1B6HBE3 V5GRP0 A0A154P6I7 A0A1B6EV42 A0A2M3ZDF1 W5JWC4 A0A2M4AYR2 A0A0A9WRN2 A0A0P4WF90 U5EWX5 A0A182FKA7 A0A2R7WZ06 A0A182YIR9 A0A2M4C115 A0A182STF4 A0A0P4VKV8 R4G8C9 A0A0V0G5I6 A0A182RJR3 A0A182JAE4 A0A2C9GRG2 A0A224XYC6 A0A182LNC5 Q7QDW9 A0A182Q4M6 A0A210QD27 A0A182W3V4 A0A023EJ00 Q17NY6 A7RFI0 R7V1T1 E0V989 A0A1W4WS63 A0A1B0CSX4 A0A1B6KAL9 A0A0K8TRA4 A0A0A9W3R7 A0A0P5GV55 A0A0P5KDY1 A0A1L8DV71 A0A2P8XVA5 E9G1G6 A0A023F7K8 V4A3J1 A0A182PSB2 A0A182NDN8 A0A336LIF4 A0A182X842 A0A182UCU3 A0A1Q3F8H1 A0A1Q3F8N7 C3YZ87 B0XCW5 A0A1J1HIU0 A0A0N7ZJC9 A0A0L8HLK3 A0A1S3D9S7 T1JL32 B3M3J9 A0A0L8HMM1 B3N2B1 A0A1D2NG19 A0A2R5LIG3 A0A0P6F091 G3VLI0 A0A293MV49 A0A2U3Z9V8 U4UIG1
Pubmed
26354079
23622113
22118469
28648823
20798317
20075255
+ More
21719571 21347285 30249741 24508170 18362917 19820115 24845553 28004739 20920257 23761445 25401762 26823975 25244985 27129103 20966253 12364791 14747013 17210077 28812685 24945155 17510324 17615350 23254933 20566863 26369729 29403074 21292972 25474469 18563158 17994087 27289101 21709235 23537049
21719571 21347285 30249741 24508170 18362917 19820115 24845553 28004739 20920257 23761445 25401762 26823975 25244985 27129103 20966253 12364791 14747013 17210077 28812685 24945155 17510324 17615350 23254933 20566863 26369729 29403074 21292972 25474469 18563158 17994087 27289101 21709235 23537049
EMBL
RSAL01000023
RVE52299.1
KQ460786
KPJ12198.1
GAIX01011059
JAA81501.1
+ More
AGBW02002847 OWR55613.1 KQ459337 KPJ01385.1 ODYU01008968 SOQ53005.1 NNAY01002114 OXU22040.1 GL438820 EFN68461.1 KQ415028 KOC58914.1 KQ982482 KYQ55852.1 KQ979608 KYN20275.1 GL888153 EGI66443.1 KQ977643 KYN00866.1 JR046287 AEY60168.1 ADTU01030036 QOIP01000003 RLU25166.1 KQ981836 KYN35051.1 KK107139 EZA57895.1 GBYB01013426 JAG83193.1 KQ435794 KOX73869.1 LBMM01007693 KMQ89505.1 NEVH01007822 PNF35312.1 KZ288189 PBC34655.1 KQ976542 KYM81141.1 KQ971338 EFA02668.2 KK852773 KDR16818.1 GEZM01044978 JAV77998.1 GEDC01014485 JAS22813.1 GECU01035675 JAS72031.1 GALX01001622 JAB66844.1 KQ434827 KZC07556.1 GECZ01028019 JAS41750.1 GGFM01005825 MBW26576.1 ADMH02000080 ETN67883.1 GGFK01012612 MBW45933.1 GBHO01036079 GBRD01009215 GDHC01017065 JAG07525.1 JAG56606.1 JAQ01564.1 GDRN01035406 JAI67403.1 GANO01000350 JAB59521.1 KK856067 PTY24769.1 GGFJ01009859 MBW59000.1 GDKW01000858 JAI55737.1 ACPB03007718 GAHY01001192 JAA76318.1 GECL01002801 JAP03323.1 APCN01005173 GFTR01003213 JAW13213.1 AAAB01008849 EAA07212.2 AXCN02002119 NEDP02004117 OWF46628.1 GAPW01004914 GEHC01000326 JAC08684.1 JAV47319.1 CH477195 EAT48412.1 DS469508 EDO49725.1 AMQN01019361 KB295646 ELU12803.1 DS234991 EEB09945.1 AJWK01026741 GEBQ01031489 GEBQ01023719 JAT08488.1 JAT16258.1 GDAI01000942 JAI16661.1 GBHO01044094 GBRD01002033 JAF99509.1 JAG63788.1 GDIQ01258530 JAJ93194.1 GDIQ01191981 GDIP01043172 LRGB01003163 JAK59744.1 JAM60543.1 KZS03920.1 GFDF01003746 JAV10338.1 PYGN01001295 PSN35933.1 GL732529 EFX86496.1 GBBI01001523 JAC17189.1 KB202591 ESO89515.1 UFQT01000020 SSX17874.1 GFDL01011180 JAV23865.1 GFDL01011158 JAV23887.1 GG666566 EEN54158.1 DS232723 EDS45160.1 CVRI01000006 CRK87968.1 GDIP01239705 JAI83696.1 KQ417858 KOF90029.1 JH431789 CH902618 EDV39260.1 KOF90030.1 CH902928 EDV44989.1 LJIJ01000051 ODN04201.1 GGLE01004991 MBY09117.1 GDIQ01056782 JAN37955.1 AEFK01120163 GFWV01019113 MAA43841.1 KB632373 ERL93794.1
AGBW02002847 OWR55613.1 KQ459337 KPJ01385.1 ODYU01008968 SOQ53005.1 NNAY01002114 OXU22040.1 GL438820 EFN68461.1 KQ415028 KOC58914.1 KQ982482 KYQ55852.1 KQ979608 KYN20275.1 GL888153 EGI66443.1 KQ977643 KYN00866.1 JR046287 AEY60168.1 ADTU01030036 QOIP01000003 RLU25166.1 KQ981836 KYN35051.1 KK107139 EZA57895.1 GBYB01013426 JAG83193.1 KQ435794 KOX73869.1 LBMM01007693 KMQ89505.1 NEVH01007822 PNF35312.1 KZ288189 PBC34655.1 KQ976542 KYM81141.1 KQ971338 EFA02668.2 KK852773 KDR16818.1 GEZM01044978 JAV77998.1 GEDC01014485 JAS22813.1 GECU01035675 JAS72031.1 GALX01001622 JAB66844.1 KQ434827 KZC07556.1 GECZ01028019 JAS41750.1 GGFM01005825 MBW26576.1 ADMH02000080 ETN67883.1 GGFK01012612 MBW45933.1 GBHO01036079 GBRD01009215 GDHC01017065 JAG07525.1 JAG56606.1 JAQ01564.1 GDRN01035406 JAI67403.1 GANO01000350 JAB59521.1 KK856067 PTY24769.1 GGFJ01009859 MBW59000.1 GDKW01000858 JAI55737.1 ACPB03007718 GAHY01001192 JAA76318.1 GECL01002801 JAP03323.1 APCN01005173 GFTR01003213 JAW13213.1 AAAB01008849 EAA07212.2 AXCN02002119 NEDP02004117 OWF46628.1 GAPW01004914 GEHC01000326 JAC08684.1 JAV47319.1 CH477195 EAT48412.1 DS469508 EDO49725.1 AMQN01019361 KB295646 ELU12803.1 DS234991 EEB09945.1 AJWK01026741 GEBQ01031489 GEBQ01023719 JAT08488.1 JAT16258.1 GDAI01000942 JAI16661.1 GBHO01044094 GBRD01002033 JAF99509.1 JAG63788.1 GDIQ01258530 JAJ93194.1 GDIQ01191981 GDIP01043172 LRGB01003163 JAK59744.1 JAM60543.1 KZS03920.1 GFDF01003746 JAV10338.1 PYGN01001295 PSN35933.1 GL732529 EFX86496.1 GBBI01001523 JAC17189.1 KB202591 ESO89515.1 UFQT01000020 SSX17874.1 GFDL01011180 JAV23865.1 GFDL01011158 JAV23887.1 GG666566 EEN54158.1 DS232723 EDS45160.1 CVRI01000006 CRK87968.1 GDIP01239705 JAI83696.1 KQ417858 KOF90029.1 JH431789 CH902618 EDV39260.1 KOF90030.1 CH902928 EDV44989.1 LJIJ01000051 ODN04201.1 GGLE01004991 MBY09117.1 GDIQ01056782 JAN37955.1 AEFK01120163 GFWV01019113 MAA43841.1 KB632373 ERL93794.1
Proteomes
UP000283053
UP000053240
UP000007151
UP000053268
UP000215335
UP000000311
+ More
UP000002358 UP000053825 UP000075809 UP000078492 UP000007755 UP000078542 UP000005205 UP000279307 UP000005203 UP000078541 UP000053097 UP000053105 UP000036403 UP000235965 UP000242457 UP000078540 UP000007266 UP000027135 UP000076502 UP000000673 UP000069272 UP000076408 UP000075901 UP000015103 UP000075900 UP000075880 UP000075840 UP000075882 UP000007062 UP000075886 UP000242188 UP000075920 UP000008820 UP000001593 UP000014760 UP000009046 UP000192223 UP000092461 UP000076858 UP000245037 UP000000305 UP000030746 UP000075885 UP000075884 UP000076407 UP000075902 UP000001554 UP000002320 UP000183832 UP000053454 UP000079169 UP000007801 UP000094527 UP000007648 UP000245340 UP000030742
UP000002358 UP000053825 UP000075809 UP000078492 UP000007755 UP000078542 UP000005205 UP000279307 UP000005203 UP000078541 UP000053097 UP000053105 UP000036403 UP000235965 UP000242457 UP000078540 UP000007266 UP000027135 UP000076502 UP000000673 UP000069272 UP000076408 UP000075901 UP000015103 UP000075900 UP000075880 UP000075840 UP000075882 UP000007062 UP000075886 UP000242188 UP000075920 UP000008820 UP000001593 UP000014760 UP000009046 UP000192223 UP000092461 UP000076858 UP000245037 UP000000305 UP000030746 UP000075885 UP000075884 UP000076407 UP000075902 UP000001554 UP000002320 UP000183832 UP000053454 UP000079169 UP000007801 UP000094527 UP000007648 UP000245340 UP000030742
Interpro
SUPFAM
SSF46785
SSF46785
Gene 3D
CDD
ProteinModelPortal
A0A3S2NYL2
A0A0N1IF32
S4NRU2
A0A212FPH1
A0A194Q8U1
A0A2H1WIV9
+ More
A0A232EUK8 E2ADU9 K7J5X7 A0A0L7QJZ3 A0A151X682 A0A195E513 F4WH77 A0A151IH42 V9IGE6 A0A158NXG2 A0A3L8DXS3 A0A088AGW9 A0A195F3T8 A0A026WPE8 A0A0C9RZ94 A0A0M9A1H3 A0A0J7NA31 A0A2J7R3A9 A0A2A3ESC0 A0A195B9D0 D2A1I8 A0A067R1V2 A0A1Y1LZ56 A0A1B6DAU7 A0A1B6HBE3 V5GRP0 A0A154P6I7 A0A1B6EV42 A0A2M3ZDF1 W5JWC4 A0A2M4AYR2 A0A0A9WRN2 A0A0P4WF90 U5EWX5 A0A182FKA7 A0A2R7WZ06 A0A182YIR9 A0A2M4C115 A0A182STF4 A0A0P4VKV8 R4G8C9 A0A0V0G5I6 A0A182RJR3 A0A182JAE4 A0A2C9GRG2 A0A224XYC6 A0A182LNC5 Q7QDW9 A0A182Q4M6 A0A210QD27 A0A182W3V4 A0A023EJ00 Q17NY6 A7RFI0 R7V1T1 E0V989 A0A1W4WS63 A0A1B0CSX4 A0A1B6KAL9 A0A0K8TRA4 A0A0A9W3R7 A0A0P5GV55 A0A0P5KDY1 A0A1L8DV71 A0A2P8XVA5 E9G1G6 A0A023F7K8 V4A3J1 A0A182PSB2 A0A182NDN8 A0A336LIF4 A0A182X842 A0A182UCU3 A0A1Q3F8H1 A0A1Q3F8N7 C3YZ87 B0XCW5 A0A1J1HIU0 A0A0N7ZJC9 A0A0L8HLK3 A0A1S3D9S7 T1JL32 B3M3J9 A0A0L8HMM1 B3N2B1 A0A1D2NG19 A0A2R5LIG3 A0A0P6F091 G3VLI0 A0A293MV49 A0A2U3Z9V8 U4UIG1
A0A232EUK8 E2ADU9 K7J5X7 A0A0L7QJZ3 A0A151X682 A0A195E513 F4WH77 A0A151IH42 V9IGE6 A0A158NXG2 A0A3L8DXS3 A0A088AGW9 A0A195F3T8 A0A026WPE8 A0A0C9RZ94 A0A0M9A1H3 A0A0J7NA31 A0A2J7R3A9 A0A2A3ESC0 A0A195B9D0 D2A1I8 A0A067R1V2 A0A1Y1LZ56 A0A1B6DAU7 A0A1B6HBE3 V5GRP0 A0A154P6I7 A0A1B6EV42 A0A2M3ZDF1 W5JWC4 A0A2M4AYR2 A0A0A9WRN2 A0A0P4WF90 U5EWX5 A0A182FKA7 A0A2R7WZ06 A0A182YIR9 A0A2M4C115 A0A182STF4 A0A0P4VKV8 R4G8C9 A0A0V0G5I6 A0A182RJR3 A0A182JAE4 A0A2C9GRG2 A0A224XYC6 A0A182LNC5 Q7QDW9 A0A182Q4M6 A0A210QD27 A0A182W3V4 A0A023EJ00 Q17NY6 A7RFI0 R7V1T1 E0V989 A0A1W4WS63 A0A1B0CSX4 A0A1B6KAL9 A0A0K8TRA4 A0A0A9W3R7 A0A0P5GV55 A0A0P5KDY1 A0A1L8DV71 A0A2P8XVA5 E9G1G6 A0A023F7K8 V4A3J1 A0A182PSB2 A0A182NDN8 A0A336LIF4 A0A182X842 A0A182UCU3 A0A1Q3F8H1 A0A1Q3F8N7 C3YZ87 B0XCW5 A0A1J1HIU0 A0A0N7ZJC9 A0A0L8HLK3 A0A1S3D9S7 T1JL32 B3M3J9 A0A0L8HMM1 B3N2B1 A0A1D2NG19 A0A2R5LIG3 A0A0P6F091 G3VLI0 A0A293MV49 A0A2U3Z9V8 U4UIG1
PDB
6B9X
E-value=2.32892e-08,
Score=135
Ontologies
GO
GO:0001919
GO:0043410
GO:0042632
GO:0016197
GO:0032008
GO:0071230
GO:0007040
GO:0071986
GO:0003700
GO:0003677
GO:0016602
GO:0034613
GO:0005764
GO:0006334
GO:0005634
GO:0000786
GO:0001558
GO:0032418
GO:0010874
GO:0060090
GO:0060620
GO:0010872
GO:0005085
GO:0007032
GO:0031902
GO:0032439
GO:0045121
GO:0016876
GO:0043039
PANTHER
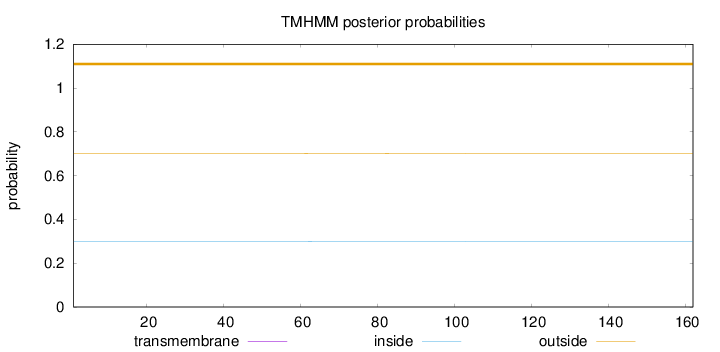
Topology
Subcellular location
Chromosome
Nucleus
Nucleus
Length:
162
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00348
Exp number, first 60 AAs:
4e-05
Total prob of N-in:
0.29975
outside
1 - 162
Population Genetic Test Statistics
Pi
235.990971
Theta
168.27078
Tajima's D
0.686746
CLR
0.811194
CSRT
0.571971401429929
Interpretation
Uncertain
