Pre Gene Modal
BGIBMGA009329
Annotation
PREDICTED:_endoplasmic_reticulum-Golgi_intermediate_compartment_protein_2_[Papilio_machaon]
Location in the cell
Cytoplasmic Reliability : 1.52 PlasmaMembrane Reliability : 1.053
Sequence
CDS
ATGTTAAGGTATCGAGGGAAAAAGAACATTTTGGAAAAAGTAAAGGAATACGATGCTTTCCCTAAAGTTCCTGATGAATACGTCGCGAGTACCCCGGTAGGAGGAACGTTTTCAGTTATCACATTTTTCATTATCCTTTGGTTAATTTACAGTGAAGTTACGTACTATTTAGACAGTAATTTAGTATTCAAATTTATGCCAGACATAGACATGGACGAAAAGTTACGAATAAACATCGATATAACCGTAGCAATGCCTTGCTCGAACATCGGTTCTGATATCTTGGACTCTACCTCGCAAAGTGTTTTCGGATTCGGTGAATTACAAGAGGAAGACACTTGGTTCGAGCTCACAGAAGAACAACAAGACGCGTTTGACGCTATAAAGTATATAAACTCTTATTTAAGAGAAGAGTATCATTCCGTTTGGCAGCTACTTTGGAAAAAGGGTCATGGCTCCGTTCGCTCTACTATACCAGAGCGTAAAACGAAAATGAACAAACGGCCGGATGCCTGTAGACTTCATGGAGTTCTGACGTTAAATAAAGTGGCTGGCAACTTCCACATAACTGCAGGAAAGAGTCTACATCTGCCCCGTGGGCACATACACTTAAATATGCTCTTTGATGATACTCCACAGAACTTCAGTCACAGGATACACCGTCTCAGTTTTGGCAGTCCAACCAATGGGATTATTTACCCTCTAGAAGGTGACGAGAAGATCACGAATGATGAAAATATGCTATATCAGTATTTTGTTGATGTTGTACCAACTGACGTTGACAATACATTTGAGACAGTAAAAGCATATCAATATTCAGTTAAAGAGTTAGAAAGACCTATAAGTCACGTGAAGGGATCTCATGGAGTTCCTGGTATTTTCTTCAAATACGATATGGCTGCATTGAAAGTTAAAGTGTATCAAGAGAGAGAAAACATGCTGCAGTTCACGTTAAGACTATTTTCAGTAATAGGAGGCATTTATGTTATAATAGGATTTATTAATGCAATAGTGTTAACGTTACAATCATACTTTGTGACAAAACCAATTAAAGATAAAAAACAGTTAGAGGTTAAAACACCTCAAAATAACTACACAACCAATGCTTTGTTTGTGGCAACAGAGTTAAGTGTTCCAGTTGAACTTGTAAAACAATAA
Protein
MLRYRGKKNILEKVKEYDAFPKVPDEYVASTPVGGTFSVITFFIILWLIYSEVTYYLDSNLVFKFMPDIDMDEKLRINIDITVAMPCSNIGSDILDSTSQSVFGFGELQEEDTWFELTEEQQDAFDAIKYINSYLREEYHSVWQLLWKKGHGSVRSTIPERKTKMNKRPDACRLHGVLTLNKVAGNFHITAGKSLHLPRGHIHLNMLFDDTPQNFSHRIHRLSFGSPTNGIIYPLEGDEKITNDENMLYQYFVDVVPTDVDNTFETVKAYQYSVKELERPISHVKGSHGVPGIFFKYDMAALKVKVYQERENMLQFTLRLFSVIGGIYVIIGFINAIVLTLQSYFVTKPIKDKKQLEVKTPQNNYTTNALFVATELSVPVELVKQ
Summary
Uniprot
H9JIH9
A0A2A4JDB7
A0A1E1WB79
A0A2H1X1W7
A0A3S2LF20
A0A0N1IGK2
+ More
A0A194QD58 A0A212FPE4 A0A0L7LS97 S4PHL9 A0A1Y1LZU7 A0A067QH48 A0A195CL78 E2A0I6 A0A0T6BAF0 A0A195D9Y7 F4WZ14 A0A195F3X3 A0A1B6HFA2 A0A195BQR0 A0A158NDL4 A0A1B6GL18 A0A154PI72 A0A1B6KXN3 A0A2M4BRP7 A0A088A2M3 A0A2A3E563 A0A182FEN4 A0A0L7RB96 A0A2M4BRB1 A0A2M3Z8X1 A0A2M4AB48 A0A0J7KIQ1 A0A151XES0 A0A2M4CSD4 A0A2M4BRF8 A0A2M4CSG9 A0A1L8DVG1 E2BHN7 E9IU73 A0A3L8E031 A0A0M9A1W8 A0A182RUW7 D6WF35 A0A182QSX9 A0A182VAK6 Q7Q9U1 A0A182HPE5 A0A182X0X8 A0A182WMA3 A0A182YRM6 A0A182UFQ6 U5EHR7 A0A1B6E230 A0A182NLV7 A0A182IWX0 A0A2P8Y317 A0A182NZR9 E0VCV5 A0A232EM79 K7J8C3 A0A336L6N7 A0A336N1E6 A0A1W4WFI3 A0A026WHN6 A0A084WKY2 T1HI76 W5JQC2 A0A0P4VQ11 A0A023F2D1 A0A0A9YSQ9 A0A146KX15 A0A0K8T9I2 J3JU94 A0A0V0G4Q2 A0A069DTM4 U4U9K2 A0A182LB54 A0A423SW94 V5GK54 A0A1D2NBW4 A0A224XEW5 A0A1W7RAJ2 A0A0K8TST3 A0A226ESL0 C1BT41 A0A0K2V8W0 E9FSB5 A0A0P5TGC1 A0A2S2QYH3 A0A0N8AN99 A0A0P5NDZ5 A0A0P6IGK1 A0A0P6AD16 A0A0P5KHN5 A0A210QDD3 A0A087TXM9 T1JCT3 A0A2R2MLP5 W8C1V5
A0A194QD58 A0A212FPE4 A0A0L7LS97 S4PHL9 A0A1Y1LZU7 A0A067QH48 A0A195CL78 E2A0I6 A0A0T6BAF0 A0A195D9Y7 F4WZ14 A0A195F3X3 A0A1B6HFA2 A0A195BQR0 A0A158NDL4 A0A1B6GL18 A0A154PI72 A0A1B6KXN3 A0A2M4BRP7 A0A088A2M3 A0A2A3E563 A0A182FEN4 A0A0L7RB96 A0A2M4BRB1 A0A2M3Z8X1 A0A2M4AB48 A0A0J7KIQ1 A0A151XES0 A0A2M4CSD4 A0A2M4BRF8 A0A2M4CSG9 A0A1L8DVG1 E2BHN7 E9IU73 A0A3L8E031 A0A0M9A1W8 A0A182RUW7 D6WF35 A0A182QSX9 A0A182VAK6 Q7Q9U1 A0A182HPE5 A0A182X0X8 A0A182WMA3 A0A182YRM6 A0A182UFQ6 U5EHR7 A0A1B6E230 A0A182NLV7 A0A182IWX0 A0A2P8Y317 A0A182NZR9 E0VCV5 A0A232EM79 K7J8C3 A0A336L6N7 A0A336N1E6 A0A1W4WFI3 A0A026WHN6 A0A084WKY2 T1HI76 W5JQC2 A0A0P4VQ11 A0A023F2D1 A0A0A9YSQ9 A0A146KX15 A0A0K8T9I2 J3JU94 A0A0V0G4Q2 A0A069DTM4 U4U9K2 A0A182LB54 A0A423SW94 V5GK54 A0A1D2NBW4 A0A224XEW5 A0A1W7RAJ2 A0A0K8TST3 A0A226ESL0 C1BT41 A0A0K2V8W0 E9FSB5 A0A0P5TGC1 A0A2S2QYH3 A0A0N8AN99 A0A0P5NDZ5 A0A0P6IGK1 A0A0P6AD16 A0A0P5KHN5 A0A210QDD3 A0A087TXM9 T1JCT3 A0A2R2MLP5 W8C1V5
Pubmed
19121390
26354079
22118469
26227816
23622113
28004739
+ More
24845553 20798317 21719571 21347285 21282665 30249741 18362917 19820115 12364791 14747013 17210077 25244985 29403074 20566863 28648823 20075255 24508170 24438588 20920257 23761445 27129103 25474469 25401762 26823975 22516182 23537049 26334808 20966253 27289101 26369729 21292972 28812685 24495485
24845553 20798317 21719571 21347285 21282665 30249741 18362917 19820115 12364791 14747013 17210077 25244985 29403074 20566863 28648823 20075255 24508170 24438588 20920257 23761445 27129103 25474469 25401762 26823975 22516182 23537049 26334808 20966253 27289101 26369729 21292972 28812685 24495485
EMBL
BABH01027194
NWSH01001966
PCG69544.1
GDQN01006933
JAT84121.1
ODYU01012826
+ More
SOQ59323.1 RSAL01000023 RVE52300.1 KQ460786 KPJ12197.1 KQ459337 KPJ01386.1 AGBW02002847 OWR55614.1 JTDY01000247 KOB78071.1 GAIX01005750 JAA86810.1 GEZM01044408 JAV78248.1 KK853443 KDR07600.1 KQ977600 KYN01470.1 GL435626 EFN72889.1 LJIG01002628 KRT84312.1 KQ981082 KYN09698.1 GL888465 EGI60517.1 KQ981820 KYN35158.1 GECU01034348 JAS73358.1 KQ976424 KYM88299.1 ADTU01012715 GECZ01006679 JAS63090.1 KQ434899 KZC11018.1 GEBQ01023787 JAT16190.1 GGFJ01006460 MBW55601.1 KZ288367 PBC26830.1 KQ414617 KOC68113.1 GGFJ01006459 MBW55600.1 GGFM01004238 MBW24989.1 GGFK01004686 MBW38007.1 LBMM01007007 KMQ90124.1 KQ982254 KYQ58768.1 GGFL01004088 MBW68266.1 GGFJ01006461 MBW55602.1 GGFL01004089 MBW68267.1 GFDF01003681 JAV10403.1 GL448322 EFN84785.1 GL765847 EFZ15869.1 QOIP01000002 RLU25823.1 KQ435793 KOX74253.1 KQ971327 EEZ99853.1 AXCN02000946 AAAB01008900 EAA09437.3 APCN01002983 GANO01002909 JAB56962.1 GEDC01029356 GEDC01005305 JAS07942.1 JAS31993.1 PYGN01000989 PSN38657.1 DS235065 EEB11211.1 NNAY01003411 OXU19456.1 AAZX01010098 UFQS01002238 UFQT01002238 SSX13566.1 SSX32993.1 UFQT01003921 SSX35369.1 KK107200 EZA55498.1 ATLV01024157 KE525350 KFB50876.1 ACPB03006711 ADMH02000413 ETN66612.1 GDKW01002704 JAI53891.1 GBBI01003441 JAC15271.1 GBHO01008958 GBHO01008957 GBHO01008956 JAG34646.1 JAG34647.1 JAG34648.1 GDHC01021429 GDHC01018464 JAP97199.1 JAQ00165.1 GBRD01004002 GBRD01004001 JAG61820.1 APGK01030369 BT126807 KB740770 AEE61769.1 ENN79004.1 GECL01003097 JAP03027.1 GBGD01001773 JAC87116.1 KB632132 ERL89008.1 QCYY01002671 ROT68398.1 GALX01004027 JAB64439.1 LJIJ01000105 ODN02575.1 GFTR01005411 JAW11015.1 GFAH01000219 JAV48170.1 GDAI01000410 JAI17193.1 LNIX01000002 OXA60208.1 BT077770 ACO12194.1 HACA01029617 CDW46978.1 GL732523 EFX90347.1 GDIP01126604 JAL77110.1 GGMS01013572 MBY82775.1 GDIQ01259067 JAJ92657.1 GDIQ01146748 JAL04978.1 GDIQ01006331 JAN88406.1 GDIP01032352 JAM71363.1 GDIQ01209570 JAK42155.1 NEDP02004093 OWF46754.1 KK117218 KFM69868.1 JH432078 GAMC01006839 JAB99716.1
SOQ59323.1 RSAL01000023 RVE52300.1 KQ460786 KPJ12197.1 KQ459337 KPJ01386.1 AGBW02002847 OWR55614.1 JTDY01000247 KOB78071.1 GAIX01005750 JAA86810.1 GEZM01044408 JAV78248.1 KK853443 KDR07600.1 KQ977600 KYN01470.1 GL435626 EFN72889.1 LJIG01002628 KRT84312.1 KQ981082 KYN09698.1 GL888465 EGI60517.1 KQ981820 KYN35158.1 GECU01034348 JAS73358.1 KQ976424 KYM88299.1 ADTU01012715 GECZ01006679 JAS63090.1 KQ434899 KZC11018.1 GEBQ01023787 JAT16190.1 GGFJ01006460 MBW55601.1 KZ288367 PBC26830.1 KQ414617 KOC68113.1 GGFJ01006459 MBW55600.1 GGFM01004238 MBW24989.1 GGFK01004686 MBW38007.1 LBMM01007007 KMQ90124.1 KQ982254 KYQ58768.1 GGFL01004088 MBW68266.1 GGFJ01006461 MBW55602.1 GGFL01004089 MBW68267.1 GFDF01003681 JAV10403.1 GL448322 EFN84785.1 GL765847 EFZ15869.1 QOIP01000002 RLU25823.1 KQ435793 KOX74253.1 KQ971327 EEZ99853.1 AXCN02000946 AAAB01008900 EAA09437.3 APCN01002983 GANO01002909 JAB56962.1 GEDC01029356 GEDC01005305 JAS07942.1 JAS31993.1 PYGN01000989 PSN38657.1 DS235065 EEB11211.1 NNAY01003411 OXU19456.1 AAZX01010098 UFQS01002238 UFQT01002238 SSX13566.1 SSX32993.1 UFQT01003921 SSX35369.1 KK107200 EZA55498.1 ATLV01024157 KE525350 KFB50876.1 ACPB03006711 ADMH02000413 ETN66612.1 GDKW01002704 JAI53891.1 GBBI01003441 JAC15271.1 GBHO01008958 GBHO01008957 GBHO01008956 JAG34646.1 JAG34647.1 JAG34648.1 GDHC01021429 GDHC01018464 JAP97199.1 JAQ00165.1 GBRD01004002 GBRD01004001 JAG61820.1 APGK01030369 BT126807 KB740770 AEE61769.1 ENN79004.1 GECL01003097 JAP03027.1 GBGD01001773 JAC87116.1 KB632132 ERL89008.1 QCYY01002671 ROT68398.1 GALX01004027 JAB64439.1 LJIJ01000105 ODN02575.1 GFTR01005411 JAW11015.1 GFAH01000219 JAV48170.1 GDAI01000410 JAI17193.1 LNIX01000002 OXA60208.1 BT077770 ACO12194.1 HACA01029617 CDW46978.1 GL732523 EFX90347.1 GDIP01126604 JAL77110.1 GGMS01013572 MBY82775.1 GDIQ01259067 JAJ92657.1 GDIQ01146748 JAL04978.1 GDIQ01006331 JAN88406.1 GDIP01032352 JAM71363.1 GDIQ01209570 JAK42155.1 NEDP02004093 OWF46754.1 KK117218 KFM69868.1 JH432078 GAMC01006839 JAB99716.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000053268
UP000007151
+ More
UP000037510 UP000027135 UP000078542 UP000000311 UP000078492 UP000007755 UP000078541 UP000078540 UP000005205 UP000076502 UP000005203 UP000242457 UP000069272 UP000053825 UP000036403 UP000075809 UP000008237 UP000279307 UP000053105 UP000075900 UP000007266 UP000075886 UP000075903 UP000007062 UP000075840 UP000076407 UP000075920 UP000076408 UP000075902 UP000075884 UP000075880 UP000245037 UP000075885 UP000009046 UP000215335 UP000002358 UP000192223 UP000053097 UP000030765 UP000015103 UP000000673 UP000019118 UP000030742 UP000075882 UP000283509 UP000094527 UP000198287 UP000000305 UP000242188 UP000054359 UP000085678
UP000037510 UP000027135 UP000078542 UP000000311 UP000078492 UP000007755 UP000078541 UP000078540 UP000005205 UP000076502 UP000005203 UP000242457 UP000069272 UP000053825 UP000036403 UP000075809 UP000008237 UP000279307 UP000053105 UP000075900 UP000007266 UP000075886 UP000075903 UP000007062 UP000075840 UP000076407 UP000075920 UP000076408 UP000075902 UP000075884 UP000075880 UP000245037 UP000075885 UP000009046 UP000215335 UP000002358 UP000192223 UP000053097 UP000030765 UP000015103 UP000000673 UP000019118 UP000030742 UP000075882 UP000283509 UP000094527 UP000198287 UP000000305 UP000242188 UP000054359 UP000085678
PRIDE
Pfam
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
H9JIH9
A0A2A4JDB7
A0A1E1WB79
A0A2H1X1W7
A0A3S2LF20
A0A0N1IGK2
+ More
A0A194QD58 A0A212FPE4 A0A0L7LS97 S4PHL9 A0A1Y1LZU7 A0A067QH48 A0A195CL78 E2A0I6 A0A0T6BAF0 A0A195D9Y7 F4WZ14 A0A195F3X3 A0A1B6HFA2 A0A195BQR0 A0A158NDL4 A0A1B6GL18 A0A154PI72 A0A1B6KXN3 A0A2M4BRP7 A0A088A2M3 A0A2A3E563 A0A182FEN4 A0A0L7RB96 A0A2M4BRB1 A0A2M3Z8X1 A0A2M4AB48 A0A0J7KIQ1 A0A151XES0 A0A2M4CSD4 A0A2M4BRF8 A0A2M4CSG9 A0A1L8DVG1 E2BHN7 E9IU73 A0A3L8E031 A0A0M9A1W8 A0A182RUW7 D6WF35 A0A182QSX9 A0A182VAK6 Q7Q9U1 A0A182HPE5 A0A182X0X8 A0A182WMA3 A0A182YRM6 A0A182UFQ6 U5EHR7 A0A1B6E230 A0A182NLV7 A0A182IWX0 A0A2P8Y317 A0A182NZR9 E0VCV5 A0A232EM79 K7J8C3 A0A336L6N7 A0A336N1E6 A0A1W4WFI3 A0A026WHN6 A0A084WKY2 T1HI76 W5JQC2 A0A0P4VQ11 A0A023F2D1 A0A0A9YSQ9 A0A146KX15 A0A0K8T9I2 J3JU94 A0A0V0G4Q2 A0A069DTM4 U4U9K2 A0A182LB54 A0A423SW94 V5GK54 A0A1D2NBW4 A0A224XEW5 A0A1W7RAJ2 A0A0K8TST3 A0A226ESL0 C1BT41 A0A0K2V8W0 E9FSB5 A0A0P5TGC1 A0A2S2QYH3 A0A0N8AN99 A0A0P5NDZ5 A0A0P6IGK1 A0A0P6AD16 A0A0P5KHN5 A0A210QDD3 A0A087TXM9 T1JCT3 A0A2R2MLP5 W8C1V5
A0A194QD58 A0A212FPE4 A0A0L7LS97 S4PHL9 A0A1Y1LZU7 A0A067QH48 A0A195CL78 E2A0I6 A0A0T6BAF0 A0A195D9Y7 F4WZ14 A0A195F3X3 A0A1B6HFA2 A0A195BQR0 A0A158NDL4 A0A1B6GL18 A0A154PI72 A0A1B6KXN3 A0A2M4BRP7 A0A088A2M3 A0A2A3E563 A0A182FEN4 A0A0L7RB96 A0A2M4BRB1 A0A2M3Z8X1 A0A2M4AB48 A0A0J7KIQ1 A0A151XES0 A0A2M4CSD4 A0A2M4BRF8 A0A2M4CSG9 A0A1L8DVG1 E2BHN7 E9IU73 A0A3L8E031 A0A0M9A1W8 A0A182RUW7 D6WF35 A0A182QSX9 A0A182VAK6 Q7Q9U1 A0A182HPE5 A0A182X0X8 A0A182WMA3 A0A182YRM6 A0A182UFQ6 U5EHR7 A0A1B6E230 A0A182NLV7 A0A182IWX0 A0A2P8Y317 A0A182NZR9 E0VCV5 A0A232EM79 K7J8C3 A0A336L6N7 A0A336N1E6 A0A1W4WFI3 A0A026WHN6 A0A084WKY2 T1HI76 W5JQC2 A0A0P4VQ11 A0A023F2D1 A0A0A9YSQ9 A0A146KX15 A0A0K8T9I2 J3JU94 A0A0V0G4Q2 A0A069DTM4 U4U9K2 A0A182LB54 A0A423SW94 V5GK54 A0A1D2NBW4 A0A224XEW5 A0A1W7RAJ2 A0A0K8TST3 A0A226ESL0 C1BT41 A0A0K2V8W0 E9FSB5 A0A0P5TGC1 A0A2S2QYH3 A0A0N8AN99 A0A0P5NDZ5 A0A0P6IGK1 A0A0P6AD16 A0A0P5KHN5 A0A210QDD3 A0A087TXM9 T1JCT3 A0A2R2MLP5 W8C1V5
PDB
3ZLC
E-value=7.14587e-07,
Score=127
Ontologies
GO
PANTHER
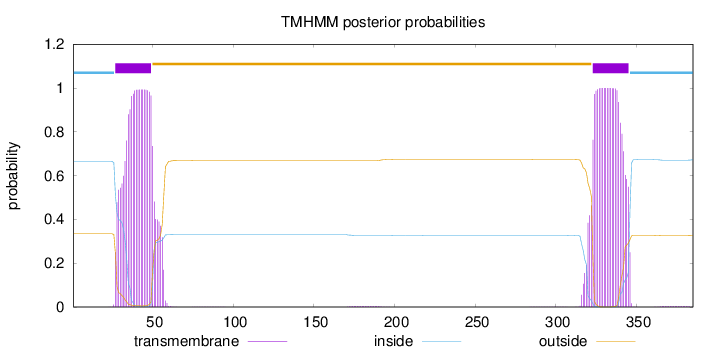
Topology
Length:
385
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
45.27371
Exp number, first 60 AAs:
22.31459
Total prob of N-in:
0.66581
POSSIBLE N-term signal
sequence
inside
1 - 26
TMhelix
27 - 49
outside
50 - 322
TMhelix
323 - 345
inside
346 - 385
Population Genetic Test Statistics
Pi
81.278261
Theta
192.228075
Tajima's D
-1.739822
CLR
791.783086
CSRT
0.031898405079746
Interpretation
Uncertain
