Gene
KWMTBOMO08524
Pre Gene Modal
BGIBMGA009333
Annotation
PREDICTED:_amyloid_beta_A4_precursor_protein-binding_family_B_member_2-like_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.487
Sequence
CDS
ATGCACGACGGCAAAACCACGCCCGACGATTACGACAACTACCTGGACAACAGACGAGTTGAAGACGAGGCTTCGTCGCCACTAAACAAGACAAATGAAAAGAACGGCTTCATAACGCCCCCACAACAGAATGGTGGTGACGACTCCCCTCCTCCGGAGAAGGAGAGGGCTGACCAGGAAGATACGGAGAAGAGTCACGCGGCACCCGTTCCCCACTCGGCTGAAGGACCCAATGATGATCTGTACGCGATACCAGTGAAGCTCAGACCGAAGAAAGAACCTCAGCTACCACCAGGATGGGAAAAACACGAGGATAACGACGGCCCATACTATTGGCACATAAAGAGCGGCACAATTCAAAGAGAGATTCCCAAAATGCCCCCCATTGAAGCTAGGGAATCCCGTATTTCCATGGTCAGAGACTGCTCGAATCTCTCCGAAGTCAGCAAATATGAGGGTGCAGTGATGACCTCGTCTGTGACCAGGAGTACAACAAGTGGAGCTCTCGACCATGTCGATCAAGAAGCTGAGAGGAAACGAAAAGAGGAGATTTCTTACAAACGCCGCAGTTACCCAGCCCGTCCAGAACCAGACAACGGGCGTGCTGTACGATTCTTCGTTCGTTCCCTCGGCTGGGTCGAGATCTCAGAGTCGGATCTGACCCCAGAACGATCCAGTAGAGCAGTCAACAAATGCATCGTGGACCTCAGCTTGGGTCGAAATGATCTTTTAGATCAAGTTGGTCGTTGGGGCGATGGTAAGGATCTGTTCATGGATCTAGATGATGGCGCCTTGAAACTGATAGATCCAGAGAGCCTGAACACACTTCACACACAGCCCATTCACACTATACGTGTTTGGGGAGTTGGAAGGGATAATGGACGAGATTTTGCATATGTGGCTCGGGATCGCGCTACTCGCAAGCACATGTGCCACGTGTTCCGATGCGAGGCTCCTGCCCGTACCATAGCCAATGCACTCCGGGACATATGCAAGCGCATCATGATTGAACGCTCCTTACAACCGCCGCCGCGTCCCACTGACTTACCCTCAACCAGGCGTCCTCGTCCATTATCAGTGGGCTGTGCGTTCCCGACTCCTATGGAAGAGCCTCGCAAGACCGTCCGAGCGTTATACCTGGGCAGCGCCGAGGTGTCCCGGGCTACCGGCATGCGCGTGCTGAATGATGCCCTAGAGCGGTTGGCCGCGGCGCGACCTCCGTCCGCGTGGCGCCCCGTGCACGTCGCCGTGGCTCCTTCCATGATCACCATCACCGAGGAGGGGGAAACGACTCCGATAGTGGAGTGCCGCGTCCGGTACCTGTCGTTCCTGGGCATAGGGCGCGACGTCCGCCGCTGCGCCTTCATCGTGCACACCGCCCAGGAGTTGTTCGTCGCGCACGCCTTCCACACCGAGCCCTCGTCGGGCGCGCTCTGCAAGACCATCGAGGCCGCCTGCAAGCTCCGGTACCAGAAGTGTCTGGACGCGCACGGCGGGGCGGCGGCGGCGGGGGCAGGCGGCGCGGCGGGGGGCGAGGGGCGCGCGTCGCTCGGCCAGGCCCTCAAGTCTCTGGTGGGCTCGCTCACGGGCCGCCGCTCCTCCTGA
Protein
MHDGKTTPDDYDNYLDNRRVEDEASSPLNKTNEKNGFITPPQQNGGDDSPPPEKERADQEDTEKSHAAPVPHSAEGPNDDLYAIPVKLRPKKEPQLPPGWEKHEDNDGPYYWHIKSGTIQREIPKMPPIEARESRISMVRDCSNLSEVSKYEGAVMTSSVTRSTTSGALDHVDQEAERKRKEEISYKRRSYPARPEPDNGRAVRFFVRSLGWVEISESDLTPERSSRAVNKCIVDLSLGRNDLLDQVGRWGDGKDLFMDLDDGALKLIDPESLNTLHTQPIHTIRVWGVGRDNGRDFAYVARDRATRKHMCHVFRCEAPARTIANALRDICKRIMIERSLQPPPRPTDLPSTRRPRPLSVGCAFPTPMEEPRKTVRALYLGSAEVSRATGMRVLNDALERLAAARPPSAWRPVHVAVAPSMITITEEGETTPIVECRVRYLSFLGIGRDVRRCAFIVHTAQELFVAHAFHTEPSSGALCKTIEAACKLRYQKCLDAHGGAAAAGAGGAAGGEGRASLGQALKSLVGSLTGRRSS
Summary
Uniprot
H9JII3
A0A2H1WH57
A0A437BPJ2
A0A2A4JPS2
A0A194QD62
A0A0N1INW0
+ More
A0A0L7KSL1 A0A1Y1M8A8 A0A1Y1M3Z0 A0A1Y1M4C0 A0A1Y1M415 A0A1Y1M4W8 A0A1Y1M9E9 A0A0T6B9K5 D2A105 A0A088AUF6 A0A310SE65 V9ICT6 A0A2P8YVC3 A0A0L7R2D3 A0A2A3ERJ5 K7IXF4 E2BZJ1 A0A151WLS5 A0A195B6C0 V9IBF6 E2A445 F4WL26 A0A0C9R655 A0A195CI61 A0A151J945 A0A1B6EF56 A0A195FBH5 A0A1B6DD44 A0A1B6CDF2 A0A1B6MUW8 A0A1B6MC25 U4UPN9 A0A0J7LA34 N6TST1 A0A158NZL6 A0A1B6IYB6 A0A2J7R4K9 A0A232FAJ2 A0A026W7W9 A0A1W4WR47 A0A154PL54 T1HF23 A0A2H8U058 J9K3V1 A0A0P4VWF5 E0VDP9 A0A146KZ07 A0A023F3E5 A0A067RHT1 A0A0A9X2N4 A0A1B0CKN9 A0A0A9YB99 A0A146LW99 Q17C69 A0A182RJQ9 A0A182W3V0 A0A1J1J6A7 A0A182LNC2 A0A1S4H412 A0A1B0DCI3 A0A182HXB6 A0A182FKB0 A0A0K8T7M1 A0A182K496 A0A0P4VYW9 A0A182NDN4 A0A182GZR8 T1J3F3 A0A0P6DQE4 A0A0P5YXE4 A0A0P5C7Q9 A0A182PSB5 A0A0P5MBA9 A0A0P5XGC3 A0A3L8D6N6 A0A162SLQ5 A0A0N8AVA2 A0A0P5WM83 A0A0P5PNL9 A0A0P6HKB0 A0A2L2YK26 A0A087TEB6 A0A0N8BL99 A0A0P5XWY2 E9GTU4 A0A0P5CHB0 A0A0P5C7Y9 A0A0P6D1T1 A0A0P5DCI7 A0A0N8EEG3 A0A0P6CUN5 A0A0P6GYB5 A0A0P5YBF6 A0A0P6BM69
A0A0L7KSL1 A0A1Y1M8A8 A0A1Y1M3Z0 A0A1Y1M4C0 A0A1Y1M415 A0A1Y1M4W8 A0A1Y1M9E9 A0A0T6B9K5 D2A105 A0A088AUF6 A0A310SE65 V9ICT6 A0A2P8YVC3 A0A0L7R2D3 A0A2A3ERJ5 K7IXF4 E2BZJ1 A0A151WLS5 A0A195B6C0 V9IBF6 E2A445 F4WL26 A0A0C9R655 A0A195CI61 A0A151J945 A0A1B6EF56 A0A195FBH5 A0A1B6DD44 A0A1B6CDF2 A0A1B6MUW8 A0A1B6MC25 U4UPN9 A0A0J7LA34 N6TST1 A0A158NZL6 A0A1B6IYB6 A0A2J7R4K9 A0A232FAJ2 A0A026W7W9 A0A1W4WR47 A0A154PL54 T1HF23 A0A2H8U058 J9K3V1 A0A0P4VWF5 E0VDP9 A0A146KZ07 A0A023F3E5 A0A067RHT1 A0A0A9X2N4 A0A1B0CKN9 A0A0A9YB99 A0A146LW99 Q17C69 A0A182RJQ9 A0A182W3V0 A0A1J1J6A7 A0A182LNC2 A0A1S4H412 A0A1B0DCI3 A0A182HXB6 A0A182FKB0 A0A0K8T7M1 A0A182K496 A0A0P4VYW9 A0A182NDN4 A0A182GZR8 T1J3F3 A0A0P6DQE4 A0A0P5YXE4 A0A0P5C7Q9 A0A182PSB5 A0A0P5MBA9 A0A0P5XGC3 A0A3L8D6N6 A0A162SLQ5 A0A0N8AVA2 A0A0P5WM83 A0A0P5PNL9 A0A0P6HKB0 A0A2L2YK26 A0A087TEB6 A0A0N8BL99 A0A0P5XWY2 E9GTU4 A0A0P5CHB0 A0A0P5C7Y9 A0A0P6D1T1 A0A0P5DCI7 A0A0N8EEG3 A0A0P6CUN5 A0A0P6GYB5 A0A0P5YBF6 A0A0P6BM69
Pubmed
EMBL
BABH01027180
BABH01027181
ODYU01008645
SOQ52405.1
RSAL01000023
RVE52302.1
+ More
NWSH01000907 PCG73584.1 KQ459337 KPJ01391.1 KQ460786 KPJ12194.1 JTDY01006330 KOB66095.1 GEZM01041218 JAV80565.1 GEZM01041219 JAV80564.1 GEZM01041220 JAV80563.1 GEZM01041221 JAV80562.1 GEZM01041222 JAV80561.1 GEZM01041217 JAV80566.1 LJIG01002922 KRT84018.1 KQ971338 EFA02585.1 KQ766603 OAD53633.1 JR038967 AEY58467.1 PYGN01000339 PSN48198.1 KQ414666 KOC64931.1 KZ288192 PBC34358.1 AAZX01004378 AAZX01004609 AAZX01007969 AAZX01007971 AAZX01015635 GL451627 EFN78889.1 KQ982944 KYQ48842.1 KQ976587 KYM79729.1 JR038968 AEY58468.1 GL436526 EFN71811.1 GL888207 EGI64989.1 GBYB01003480 JAG73247.1 KQ977720 KYN00421.1 KQ979471 KYN21430.1 GEDC01000731 JAS36567.1 KQ981685 KYN37980.1 GEDC01013738 JAS23560.1 GEDC01025837 JAS11461.1 GEBQ01000243 JAT39734.1 GEBQ01006491 JAT33486.1 KB632390 ERL94453.1 LBMM01000153 KMR04708.1 APGK01055826 KB741270 ENN71401.1 ADTU01004729 ADTU01004730 ADTU01004731 ADTU01004732 ADTU01004733 ADTU01004734 ADTU01004735 ADTU01004736 ADTU01004737 ADTU01004738 GECU01015810 JAS91896.1 NEVH01007407 PNF35759.1 NNAY01000600 OXU27458.1 KK107353 EZA52093.1 KQ434954 KZC12582.1 ACPB03015449 GFXV01006973 MBW18778.1 ABLF02035159 GDKW01000144 JAI56451.1 DS235083 EEB11505.1 GDHC01017854 JAQ00775.1 GBBI01002946 JAC15766.1 KK852629 KDR19882.1 GBHO01041654 GBHO01029681 GBHO01029679 GBHO01029678 JAG01950.1 JAG13923.1 JAG13925.1 JAG13926.1 AJWK01016488 AJWK01016489 GBHO01017538 GBHO01014708 JAG26066.1 JAG28896.1 GDHC01008231 GDHC01006742 GDHC01006681 JAQ10398.1 JAQ11887.1 JAQ11948.1 CH477311 EAT43922.1 CVRI01000069 CRL07006.1 AAAB01008849 AJVK01005111 AJVK01005112 AJVK01005113 APCN01005173 GBRD01004651 JAG61170.1 GDRN01097116 JAI59201.1 JXUM01100173 JXUM01100174 JXUM01100175 JXUM01100176 JXUM01100177 JXUM01100178 JXUM01100179 JXUM01100180 JXUM01100181 JXUM01100182 JH431826 GDIQ01084190 GDIQ01054153 JAN10547.1 GDIP01053258 JAM50457.1 GDIP01174269 JAJ49133.1 GDIQ01166668 JAK85057.1 GDIP01074149 JAM29566.1 QOIP01000012 RLU15916.1 LRGB01000024 KZS21425.1 GDIQ01239443 GDIQ01036903 JAK12282.1 GDIP01084382 JAM19333.1 GDIQ01146741 JAL04985.1 GDIQ01018578 JAN76159.1 IAAA01032182 LAA08473.1 KK114834 KFM63455.1 GDIQ01166667 JAK85058.1 GDIP01066053 JAM37662.1 GL732564 EFX77134.1 GDIP01174267 JAJ49135.1 GDIP01174270 JAJ49132.1 GDIQ01084189 JAN10548.1 GDIP01174268 JAJ49134.1 GDIQ01034764 JAN59973.1 GDIQ01087495 JAN07242.1 GDIQ01039304 JAN55433.1 GDIP01074148 JAM29567.1 GDIP01012665 JAM91050.1
NWSH01000907 PCG73584.1 KQ459337 KPJ01391.1 KQ460786 KPJ12194.1 JTDY01006330 KOB66095.1 GEZM01041218 JAV80565.1 GEZM01041219 JAV80564.1 GEZM01041220 JAV80563.1 GEZM01041221 JAV80562.1 GEZM01041222 JAV80561.1 GEZM01041217 JAV80566.1 LJIG01002922 KRT84018.1 KQ971338 EFA02585.1 KQ766603 OAD53633.1 JR038967 AEY58467.1 PYGN01000339 PSN48198.1 KQ414666 KOC64931.1 KZ288192 PBC34358.1 AAZX01004378 AAZX01004609 AAZX01007969 AAZX01007971 AAZX01015635 GL451627 EFN78889.1 KQ982944 KYQ48842.1 KQ976587 KYM79729.1 JR038968 AEY58468.1 GL436526 EFN71811.1 GL888207 EGI64989.1 GBYB01003480 JAG73247.1 KQ977720 KYN00421.1 KQ979471 KYN21430.1 GEDC01000731 JAS36567.1 KQ981685 KYN37980.1 GEDC01013738 JAS23560.1 GEDC01025837 JAS11461.1 GEBQ01000243 JAT39734.1 GEBQ01006491 JAT33486.1 KB632390 ERL94453.1 LBMM01000153 KMR04708.1 APGK01055826 KB741270 ENN71401.1 ADTU01004729 ADTU01004730 ADTU01004731 ADTU01004732 ADTU01004733 ADTU01004734 ADTU01004735 ADTU01004736 ADTU01004737 ADTU01004738 GECU01015810 JAS91896.1 NEVH01007407 PNF35759.1 NNAY01000600 OXU27458.1 KK107353 EZA52093.1 KQ434954 KZC12582.1 ACPB03015449 GFXV01006973 MBW18778.1 ABLF02035159 GDKW01000144 JAI56451.1 DS235083 EEB11505.1 GDHC01017854 JAQ00775.1 GBBI01002946 JAC15766.1 KK852629 KDR19882.1 GBHO01041654 GBHO01029681 GBHO01029679 GBHO01029678 JAG01950.1 JAG13923.1 JAG13925.1 JAG13926.1 AJWK01016488 AJWK01016489 GBHO01017538 GBHO01014708 JAG26066.1 JAG28896.1 GDHC01008231 GDHC01006742 GDHC01006681 JAQ10398.1 JAQ11887.1 JAQ11948.1 CH477311 EAT43922.1 CVRI01000069 CRL07006.1 AAAB01008849 AJVK01005111 AJVK01005112 AJVK01005113 APCN01005173 GBRD01004651 JAG61170.1 GDRN01097116 JAI59201.1 JXUM01100173 JXUM01100174 JXUM01100175 JXUM01100176 JXUM01100177 JXUM01100178 JXUM01100179 JXUM01100180 JXUM01100181 JXUM01100182 JH431826 GDIQ01084190 GDIQ01054153 JAN10547.1 GDIP01053258 JAM50457.1 GDIP01174269 JAJ49133.1 GDIQ01166668 JAK85057.1 GDIP01074149 JAM29566.1 QOIP01000012 RLU15916.1 LRGB01000024 KZS21425.1 GDIQ01239443 GDIQ01036903 JAK12282.1 GDIP01084382 JAM19333.1 GDIQ01146741 JAL04985.1 GDIQ01018578 JAN76159.1 IAAA01032182 LAA08473.1 KK114834 KFM63455.1 GDIQ01166667 JAK85058.1 GDIP01066053 JAM37662.1 GL732564 EFX77134.1 GDIP01174267 JAJ49135.1 GDIP01174270 JAJ49132.1 GDIQ01084189 JAN10548.1 GDIP01174268 JAJ49134.1 GDIQ01034764 JAN59973.1 GDIQ01087495 JAN07242.1 GDIQ01039304 JAN55433.1 GDIP01074148 JAM29567.1 GDIP01012665 JAM91050.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000053240
UP000037510
+ More
UP000007266 UP000005203 UP000245037 UP000053825 UP000242457 UP000002358 UP000008237 UP000075809 UP000078540 UP000000311 UP000007755 UP000078542 UP000078492 UP000078541 UP000030742 UP000036403 UP000019118 UP000005205 UP000235965 UP000215335 UP000053097 UP000192223 UP000076502 UP000015103 UP000007819 UP000009046 UP000027135 UP000092461 UP000008820 UP000075900 UP000075920 UP000183832 UP000075882 UP000092462 UP000075840 UP000069272 UP000075881 UP000075884 UP000069940 UP000075885 UP000279307 UP000076858 UP000054359 UP000000305
UP000007266 UP000005203 UP000245037 UP000053825 UP000242457 UP000002358 UP000008237 UP000075809 UP000078540 UP000000311 UP000007755 UP000078542 UP000078492 UP000078541 UP000030742 UP000036403 UP000019118 UP000005205 UP000235965 UP000215335 UP000053097 UP000192223 UP000076502 UP000015103 UP000007819 UP000009046 UP000027135 UP000092461 UP000008820 UP000075900 UP000075920 UP000183832 UP000075882 UP000092462 UP000075840 UP000069272 UP000075881 UP000075884 UP000069940 UP000075885 UP000279307 UP000076858 UP000054359 UP000000305
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JII3
A0A2H1WH57
A0A437BPJ2
A0A2A4JPS2
A0A194QD62
A0A0N1INW0
+ More
A0A0L7KSL1 A0A1Y1M8A8 A0A1Y1M3Z0 A0A1Y1M4C0 A0A1Y1M415 A0A1Y1M4W8 A0A1Y1M9E9 A0A0T6B9K5 D2A105 A0A088AUF6 A0A310SE65 V9ICT6 A0A2P8YVC3 A0A0L7R2D3 A0A2A3ERJ5 K7IXF4 E2BZJ1 A0A151WLS5 A0A195B6C0 V9IBF6 E2A445 F4WL26 A0A0C9R655 A0A195CI61 A0A151J945 A0A1B6EF56 A0A195FBH5 A0A1B6DD44 A0A1B6CDF2 A0A1B6MUW8 A0A1B6MC25 U4UPN9 A0A0J7LA34 N6TST1 A0A158NZL6 A0A1B6IYB6 A0A2J7R4K9 A0A232FAJ2 A0A026W7W9 A0A1W4WR47 A0A154PL54 T1HF23 A0A2H8U058 J9K3V1 A0A0P4VWF5 E0VDP9 A0A146KZ07 A0A023F3E5 A0A067RHT1 A0A0A9X2N4 A0A1B0CKN9 A0A0A9YB99 A0A146LW99 Q17C69 A0A182RJQ9 A0A182W3V0 A0A1J1J6A7 A0A182LNC2 A0A1S4H412 A0A1B0DCI3 A0A182HXB6 A0A182FKB0 A0A0K8T7M1 A0A182K496 A0A0P4VYW9 A0A182NDN4 A0A182GZR8 T1J3F3 A0A0P6DQE4 A0A0P5YXE4 A0A0P5C7Q9 A0A182PSB5 A0A0P5MBA9 A0A0P5XGC3 A0A3L8D6N6 A0A162SLQ5 A0A0N8AVA2 A0A0P5WM83 A0A0P5PNL9 A0A0P6HKB0 A0A2L2YK26 A0A087TEB6 A0A0N8BL99 A0A0P5XWY2 E9GTU4 A0A0P5CHB0 A0A0P5C7Y9 A0A0P6D1T1 A0A0P5DCI7 A0A0N8EEG3 A0A0P6CUN5 A0A0P6GYB5 A0A0P5YBF6 A0A0P6BM69
A0A0L7KSL1 A0A1Y1M8A8 A0A1Y1M3Z0 A0A1Y1M4C0 A0A1Y1M415 A0A1Y1M4W8 A0A1Y1M9E9 A0A0T6B9K5 D2A105 A0A088AUF6 A0A310SE65 V9ICT6 A0A2P8YVC3 A0A0L7R2D3 A0A2A3ERJ5 K7IXF4 E2BZJ1 A0A151WLS5 A0A195B6C0 V9IBF6 E2A445 F4WL26 A0A0C9R655 A0A195CI61 A0A151J945 A0A1B6EF56 A0A195FBH5 A0A1B6DD44 A0A1B6CDF2 A0A1B6MUW8 A0A1B6MC25 U4UPN9 A0A0J7LA34 N6TST1 A0A158NZL6 A0A1B6IYB6 A0A2J7R4K9 A0A232FAJ2 A0A026W7W9 A0A1W4WR47 A0A154PL54 T1HF23 A0A2H8U058 J9K3V1 A0A0P4VWF5 E0VDP9 A0A146KZ07 A0A023F3E5 A0A067RHT1 A0A0A9X2N4 A0A1B0CKN9 A0A0A9YB99 A0A146LW99 Q17C69 A0A182RJQ9 A0A182W3V0 A0A1J1J6A7 A0A182LNC2 A0A1S4H412 A0A1B0DCI3 A0A182HXB6 A0A182FKB0 A0A0K8T7M1 A0A182K496 A0A0P4VYW9 A0A182NDN4 A0A182GZR8 T1J3F3 A0A0P6DQE4 A0A0P5YXE4 A0A0P5C7Q9 A0A182PSB5 A0A0P5MBA9 A0A0P5XGC3 A0A3L8D6N6 A0A162SLQ5 A0A0N8AVA2 A0A0P5WM83 A0A0P5PNL9 A0A0P6HKB0 A0A2L2YK26 A0A087TEB6 A0A0N8BL99 A0A0P5XWY2 E9GTU4 A0A0P5CHB0 A0A0P5C7Y9 A0A0P6D1T1 A0A0P5DCI7 A0A0N8EEG3 A0A0P6CUN5 A0A0P6GYB5 A0A0P5YBF6 A0A0P6BM69
PDB
3D8F
E-value=2.9333e-41,
Score=425
Ontologies
GO
PANTHER
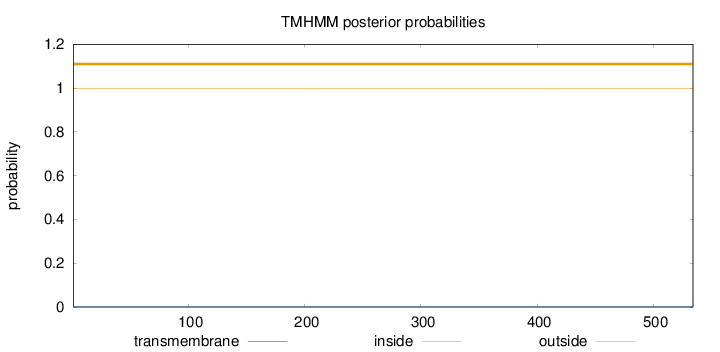
Topology
Length:
534
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00213
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00300
outside
1 - 534
Population Genetic Test Statistics
Pi
204.581191
Theta
176.947502
Tajima's D
1.025881
CLR
0.58425
CSRT
0.669066546672666
Interpretation
Uncertain
