Gene
KWMTBOMO08517
Annotation
odorant-binding_protein_3_precursor_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.508
Sequence
CDS
ATGTTCTACCCGTTTCGATTCACGCTCTTGTTTTATGGTCTCTTCGTAATTTACTTGGTTCGGGCCGAGCCGGAGAAAGAAAACCACTTCACTTTGGCTCTAAAGAAGACGCTCTTCAGCACAGCGCGCTCTTGTATGAGCCATGTCAATGCCAACGAAACTGACCTAGAATATTTGAGGAAAGACCCGCCATTTCCTGACAAAGCAGCATGCATCATAAAATGTCTTTTAGAGAAGATCGGTGTCGTGAAAAACAACAAGTACTCGAAAATGGGCTTCCTGACGGCGGTATCGCCACTCGTCTTTACAAACAAAAAGAAATTGGACCACTACAAGAGCGTATCCGAAAATTGCGAAAAGGAGATCAACCACGACCAGACAACGGAATGCGAACTTGGAAACGAGGTGGTTTCCTGCATCTTCAAATACGCACCAGAGCTGCATTTCAAAACATGA
Protein
MFYPFRFTLLFYGLFVIYLVRAEPEKENHFTLALKKTLFSTARSCMSHVNANETDLEYLRKDPPFPDKAACIIKCLLEKIGVVKNNKYSKMGFLTAVSPLVFTNKKKLDHYKSVSENCEKEINHDQTTECELGNEVVSCIFKYAPELHFKT
Summary
Uniprot
B8ZWK3
A0A2W1BAR4
H9JIL6
A0A3S2TQG4
A0A194Q7M8
A0A212EWA9
+ More
S5TNS4 A0A2H4ZB14 U5ERI6 A0A0C5BA58 A0A1J0KKM6 D7ELE9 A0A1I9HZP8 A0A385IUN6 A0A310SCZ5 A0A385IUN7 A0A1W6QYT9 R4G8H8 A0A2S0X9J7 A0A162X6J9 A0A385IUP1 A0A182JNI0 B0XC79 B0XC80 A0A2Z5EM84 A0A0R8PDK1 G0YYB8 A0A0E3U2J1 A0A1Q3FR49 A0A1J1IA71 A0A125RYU8 A0A343WGX4 A0A0R8P0J5 A0A1W6QYM8 A0A2S0X9F9 A0A182XV40 A0A0U3TUZ9 A0A1Q3FRX2 A0A084VE38 A0A182VEI6 A0A182R4S2 A0A182VRN5 B0WU11 A0A1L2JGQ7 A0A1J1ICB1 B0WLZ9 A0A142FHA1 A0A182RUA8 B0XK47 A0A096W1L6 A0A224XNA0 G5CBZ8 B0WM00 A0A142FH85 A0A142FH89 A0A182P640 A0A142K321 A0A0U3TWR6 B0WL17 Q7YSZ4 A0A385IUP5 A0A3G2YV03 A0A182UCX7 A0A182XHI4 Q8I8T3 A0A182HTW2 A0A1N7TMV4 R4FMR9 A0A109UNP7 A0A385IUM9 R4G8J5 A0A1Y1LWG4 A0A182MF96 B0WM01 A0A067XN83 F4Y5P7 G5CC00 A0A2S0X9G7 A0A2S0X9G9 A0A182N543 A0A0G3ZAP4 A0A1B6E1W8 A0A0U3BFY8 A0A182QN26 A0A0X8DCA0 A0A0K8TJD2
S5TNS4 A0A2H4ZB14 U5ERI6 A0A0C5BA58 A0A1J0KKM6 D7ELE9 A0A1I9HZP8 A0A385IUN6 A0A310SCZ5 A0A385IUN7 A0A1W6QYT9 R4G8H8 A0A2S0X9J7 A0A162X6J9 A0A385IUP1 A0A182JNI0 B0XC79 B0XC80 A0A2Z5EM84 A0A0R8PDK1 G0YYB8 A0A0E3U2J1 A0A1Q3FR49 A0A1J1IA71 A0A125RYU8 A0A343WGX4 A0A0R8P0J5 A0A1W6QYM8 A0A2S0X9F9 A0A182XV40 A0A0U3TUZ9 A0A1Q3FRX2 A0A084VE38 A0A182VEI6 A0A182R4S2 A0A182VRN5 B0WU11 A0A1L2JGQ7 A0A1J1ICB1 B0WLZ9 A0A142FHA1 A0A182RUA8 B0XK47 A0A096W1L6 A0A224XNA0 G5CBZ8 B0WM00 A0A142FH85 A0A142FH89 A0A182P640 A0A142K321 A0A0U3TWR6 B0WL17 Q7YSZ4 A0A385IUP5 A0A3G2YV03 A0A182UCX7 A0A182XHI4 Q8I8T3 A0A182HTW2 A0A1N7TMV4 R4FMR9 A0A109UNP7 A0A385IUM9 R4G8J5 A0A1Y1LWG4 A0A182MF96 B0WM01 A0A067XN83 F4Y5P7 G5CC00 A0A2S0X9G7 A0A2S0X9G9 A0A182N543 A0A0G3ZAP4 A0A1B6E1W8 A0A0U3BFY8 A0A182QN26 A0A0X8DCA0 A0A0K8TJD2
Pubmed
EMBL
FM876236
CAS90127.1
KZ150185
PZC72462.1
BABH01027137
RSAL01000023
+ More
RVE52306.1 RVE52309.1 KQ459337 KPJ01394.1 AGBW02012035 OWR45766.1 KC887507 AGS36743.1 MF975375 AUF72951.1 GANO01003722 JAB56149.1 KP071915 AJM71475.1 KX298769 APC94286.1 DS497846 EFA12066.1 KF984183 AIX97066.1 MH230201 AXY87860.1 GGOB01000076 MCH29472.1 MH230205 AXY87864.1 KY056625 ARO46432.1 ACPB03017963 ACPB03017964 GAHY01000947 JAA76563.1 MG544165 AWC08456.1 LT555354 SAJ59039.1 MH230207 AXY87866.1 DS232677 EDS44685.1 EDS44686.1 MF593912 AXB87330.1 KP082930 AKW47222.1 HQ845077 AEK32394.1 KM251646 AKC58527.1 GFDL01005099 JAV29946.1 CVRI01000047 CRK97183.1 KT358500 AMD82868.1 MF417511 AWC67998.1 KP082905 AKW47197.1 KY056626 ARO46433.1 MG544123 AWC08414.1 KU133776 ALV87601.1 GFDL01004706 JAV30339.1 ATLV01012227 KE524773 KFB36232.1 DS232098 EDS34725.1 KT875756 AOV87037.1 CRK97188.1 DS231993 EDS30813.1 KT281942 AMQ76487.1 DS233706 EDS31193.1 KC516750 AGZ04925.1 GFTR01002461 JAW13965.1 JN012600 AEQ19907.1 EDS30814.1 KT281926 AMQ76471.1 KT281930 AMQ76475.1 KT861420 AMR98356.1 KU133772 ALV87597.1 DS231979 EDS30180.1 ACPB03000296 AY340268 AAQ20833.1 MH230209 AXY87868.1 MG820724 AYP30816.1 AY146718 AAAB01008859 AAO12078.1 EAA07997.2 APCN01000586 KT875763 AOV87044.1 GAHY01000863 JAA76647.1 KT596722 AMD02857.1 MH230206 AXY87865.1 GAHY01000862 JAA76648.1 GEZM01047984 JAV76670.1 AXCM01004142 EDS30815.1 KC663686 AGO81740.1 GQ477030 ACZ58080.1 JN012602 AEQ19909.1 MG544135 AWC08426.1 MG544136 AWC08427.1 KP403731 AKM52550.2 GEDC01005399 JAS31899.1 KT251208 ALV87637.1 AXCN02000175 KT381640 AMA98131.1 GBRD01000212 JAG65609.1
RVE52306.1 RVE52309.1 KQ459337 KPJ01394.1 AGBW02012035 OWR45766.1 KC887507 AGS36743.1 MF975375 AUF72951.1 GANO01003722 JAB56149.1 KP071915 AJM71475.1 KX298769 APC94286.1 DS497846 EFA12066.1 KF984183 AIX97066.1 MH230201 AXY87860.1 GGOB01000076 MCH29472.1 MH230205 AXY87864.1 KY056625 ARO46432.1 ACPB03017963 ACPB03017964 GAHY01000947 JAA76563.1 MG544165 AWC08456.1 LT555354 SAJ59039.1 MH230207 AXY87866.1 DS232677 EDS44685.1 EDS44686.1 MF593912 AXB87330.1 KP082930 AKW47222.1 HQ845077 AEK32394.1 KM251646 AKC58527.1 GFDL01005099 JAV29946.1 CVRI01000047 CRK97183.1 KT358500 AMD82868.1 MF417511 AWC67998.1 KP082905 AKW47197.1 KY056626 ARO46433.1 MG544123 AWC08414.1 KU133776 ALV87601.1 GFDL01004706 JAV30339.1 ATLV01012227 KE524773 KFB36232.1 DS232098 EDS34725.1 KT875756 AOV87037.1 CRK97188.1 DS231993 EDS30813.1 KT281942 AMQ76487.1 DS233706 EDS31193.1 KC516750 AGZ04925.1 GFTR01002461 JAW13965.1 JN012600 AEQ19907.1 EDS30814.1 KT281926 AMQ76471.1 KT281930 AMQ76475.1 KT861420 AMR98356.1 KU133772 ALV87597.1 DS231979 EDS30180.1 ACPB03000296 AY340268 AAQ20833.1 MH230209 AXY87868.1 MG820724 AYP30816.1 AY146718 AAAB01008859 AAO12078.1 EAA07997.2 APCN01000586 KT875763 AOV87044.1 GAHY01000863 JAA76647.1 KT596722 AMD02857.1 MH230206 AXY87865.1 GAHY01000862 JAA76648.1 GEZM01047984 JAV76670.1 AXCM01004142 EDS30815.1 KC663686 AGO81740.1 GQ477030 ACZ58080.1 JN012602 AEQ19909.1 MG544135 AWC08426.1 MG544136 AWC08427.1 KP403731 AKM52550.2 GEDC01005399 JAS31899.1 KT251208 ALV87637.1 AXCN02000175 KT381640 AMA98131.1 GBRD01000212 JAG65609.1
Proteomes
Pfam
PF01395 PBP_GOBP
SUPFAM
SSF47565
SSF47565
Gene 3D
ProteinModelPortal
B8ZWK3
A0A2W1BAR4
H9JIL6
A0A3S2TQG4
A0A194Q7M8
A0A212EWA9
+ More
S5TNS4 A0A2H4ZB14 U5ERI6 A0A0C5BA58 A0A1J0KKM6 D7ELE9 A0A1I9HZP8 A0A385IUN6 A0A310SCZ5 A0A385IUN7 A0A1W6QYT9 R4G8H8 A0A2S0X9J7 A0A162X6J9 A0A385IUP1 A0A182JNI0 B0XC79 B0XC80 A0A2Z5EM84 A0A0R8PDK1 G0YYB8 A0A0E3U2J1 A0A1Q3FR49 A0A1J1IA71 A0A125RYU8 A0A343WGX4 A0A0R8P0J5 A0A1W6QYM8 A0A2S0X9F9 A0A182XV40 A0A0U3TUZ9 A0A1Q3FRX2 A0A084VE38 A0A182VEI6 A0A182R4S2 A0A182VRN5 B0WU11 A0A1L2JGQ7 A0A1J1ICB1 B0WLZ9 A0A142FHA1 A0A182RUA8 B0XK47 A0A096W1L6 A0A224XNA0 G5CBZ8 B0WM00 A0A142FH85 A0A142FH89 A0A182P640 A0A142K321 A0A0U3TWR6 B0WL17 Q7YSZ4 A0A385IUP5 A0A3G2YV03 A0A182UCX7 A0A182XHI4 Q8I8T3 A0A182HTW2 A0A1N7TMV4 R4FMR9 A0A109UNP7 A0A385IUM9 R4G8J5 A0A1Y1LWG4 A0A182MF96 B0WM01 A0A067XN83 F4Y5P7 G5CC00 A0A2S0X9G7 A0A2S0X9G9 A0A182N543 A0A0G3ZAP4 A0A1B6E1W8 A0A0U3BFY8 A0A182QN26 A0A0X8DCA0 A0A0K8TJD2
S5TNS4 A0A2H4ZB14 U5ERI6 A0A0C5BA58 A0A1J0KKM6 D7ELE9 A0A1I9HZP8 A0A385IUN6 A0A310SCZ5 A0A385IUN7 A0A1W6QYT9 R4G8H8 A0A2S0X9J7 A0A162X6J9 A0A385IUP1 A0A182JNI0 B0XC79 B0XC80 A0A2Z5EM84 A0A0R8PDK1 G0YYB8 A0A0E3U2J1 A0A1Q3FR49 A0A1J1IA71 A0A125RYU8 A0A343WGX4 A0A0R8P0J5 A0A1W6QYM8 A0A2S0X9F9 A0A182XV40 A0A0U3TUZ9 A0A1Q3FRX2 A0A084VE38 A0A182VEI6 A0A182R4S2 A0A182VRN5 B0WU11 A0A1L2JGQ7 A0A1J1ICB1 B0WLZ9 A0A142FHA1 A0A182RUA8 B0XK47 A0A096W1L6 A0A224XNA0 G5CBZ8 B0WM00 A0A142FH85 A0A142FH89 A0A182P640 A0A142K321 A0A0U3TWR6 B0WL17 Q7YSZ4 A0A385IUP5 A0A3G2YV03 A0A182UCX7 A0A182XHI4 Q8I8T3 A0A182HTW2 A0A1N7TMV4 R4FMR9 A0A109UNP7 A0A385IUM9 R4G8J5 A0A1Y1LWG4 A0A182MF96 B0WM01 A0A067XN83 F4Y5P7 G5CC00 A0A2S0X9G7 A0A2S0X9G9 A0A182N543 A0A0G3ZAP4 A0A1B6E1W8 A0A0U3BFY8 A0A182QN26 A0A0X8DCA0 A0A0K8TJD2
PDB
1P28
E-value=0.000649104,
Score=96
Ontologies
GO
Topology
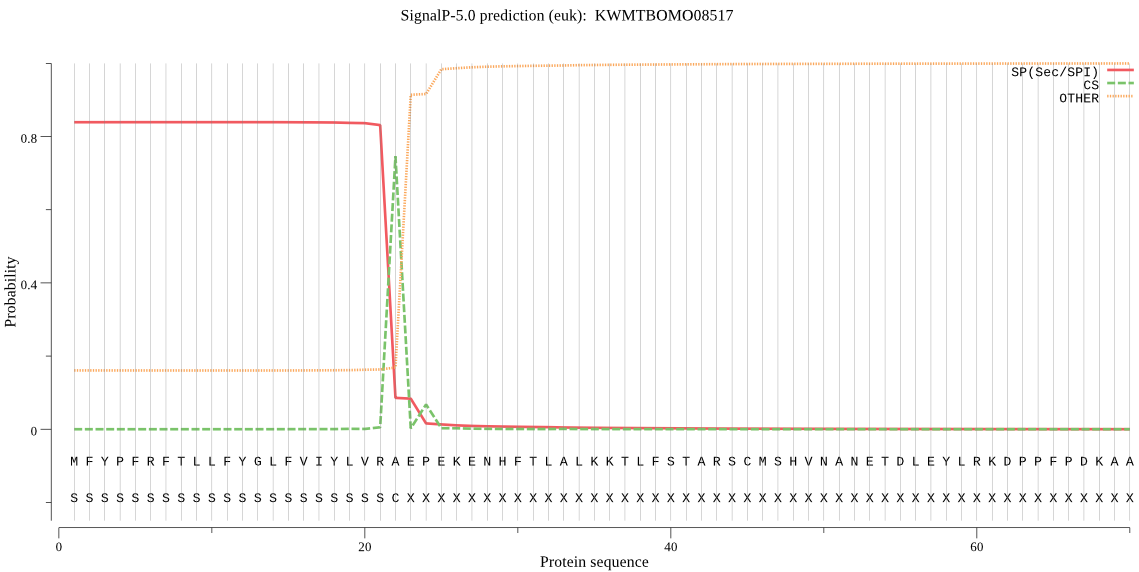
SignalP
Position: 1 - 22,
Likelihood: 0.839077
Length:
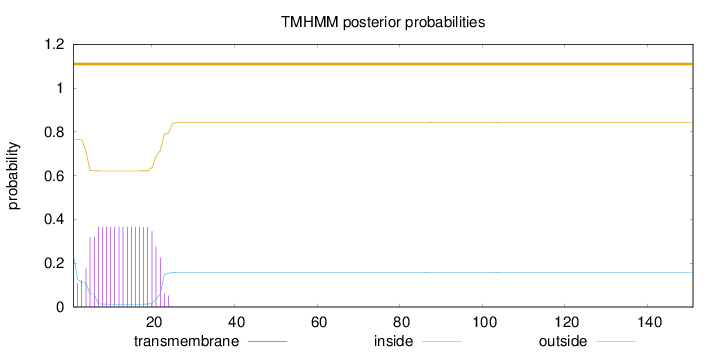
151
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
6.77942
Exp number, first 60 AAs:
6.76381
Total prob of N-in:
0.23446
outside
1 - 151
Population Genetic Test Statistics
Pi
201.790468
Theta
199.186306
Tajima's D
0.010102
CLR
0.763559
CSRT
0.372131393430328
Interpretation
Uncertain
