Pre Gene Modal
BGIBMGA009336
Annotation
PREDICTED:_leucine-rich_repeat-containing_protein_24-like_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 2.473
Sequence
CDS
ATGGGCATTCGAGTATGCGTTATTTGGTTGCTTGTGCTCATGACCATCGCCCCGACCGGAAGCGATTGGCTGACCTGTGGCAACATACCAGGATGCAACTGCAAATGGTCCTCGGGAAAGAAAACGGCATCATGCTCCTCGCTGGGATTAAAAACACCTCCAGAATTGGAATCGGACATTCAAGTTCTTGACTTGCACGGGAATCCACTCGAAACATTAGGAAAAGATGCTTTCCTGAGAGTGAATATGATAAACCTACAAAAGGTCAATCTGAGCTCGACATGTTTACGGACACTGCATTCAAACGCATTCAATCACCTTCGGATATTAATTGAATTAGATCTATCCCAGAACGAACTGACTCAGCTATCCCCTGACCTATTCAAGGCCACAGAACGTTTGAGAGTCCTAATACTTAACGATAACCCTCTAACGACGCTCGTCGCCGAGCAATTTCCTCCGTTGAGATATTTGAAGAAACTTGAAATTTCACGTTGTAAACTTCGCTCAGTGAGCTCGTTCGCTTTCACGAATTTAAGAGCATTGGAATCGGTCAATATCCAACAAAATTATCTAACGCACCTGCATCAAAGTACTTTCAATCTACCCCTCATGAATACTCTCTCCCTGTCGGATAATCCTTGGTATTGTGATTGTGGACTGCGCGAATTCCACGACTGGTTCGTGAGTAGTAATTTGGGTAATGAAGAAGTTTTATGTCTCGGTCCGGAAAGTAAAGCAAATATTTCATGGAGTGCTCTGAAGCGTGAAGATTTCTTCTGTTCACCATCTGTACTTACTTCACCTACCGTCATAAGAACTGAAGTGGGAGCAGACATCAGCTTTGGGTGTTTTGTTCGCGGTGATCCCAAGCCCGTGGTAACTTGGTCACTTAATCGTGCTGATGTATTTAACAACATTGAAAACGGTGAAATCACTATTCATAGATACGATATAGATAATTTTAATGAGTACACAGACTATTACTTAAACAAAAGTTCACAGTGGGTTAACATAAGTATAAGTAATATTACCAACGATTTAGCCGGTGAATGGAAGTGTAGTGCCAAAAGTTCTGTAGGAGATGCTTCAACCTACTTGACTATTCTTTTACCTAAAGCTAGGACAGCTAGAACAGCGACTGCTAGGGTAGTACCCGACTATTCTGCATTTTTCCTGATCGCGGGGTCCATGTTAGCTATGACAGGAGTAGGATTTGTATCGGCGTATATCTGCTGGAAGACTAGACGCCGAAGAATCCCACCTAGCAGAAGTTTCACTGACCAAGAGAAGAAACTTTTAGATTCCTCATTGGCTGCAAGCTGTGACAGAGCAAGTGGAGATATGAGCTTGTCTTATGGATTTGAAATGTTAGATAGATCCATGTCTTTGGACAGCGAAGACAATAGATGCTTAGAACCGGTCCAAATTACAGTAGAAGGTCCACCGGGATCCTTCCCTCCCCCGCCTGCGGAGTTCGCCTTGCCGGCACCATATGGGAATATTTTTATATCAGTCCAAGTTTCTGGTCACAACGATGAATACCCTGATTTATTGGGTGGTGGAACAACATTACCTAGGCGGAATACGACATGTTTCTTGAAGTCTACTTATGATAATATGGGTCCTAGGATTACGGCAGCCGGTAGTTCGACGTGGTCTTTGCCGGGTGCTAATGACAAAAATACTGTCAAAGAGAATCTCTTGACGACGCCTCTGTCTAACTTTTCTACTGAATTTACCGCTTTGTAA
Protein
MGIRVCVIWLLVLMTIAPTGSDWLTCGNIPGCNCKWSSGKKTASCSSLGLKTPPELESDIQVLDLHGNPLETLGKDAFLRVNMINLQKVNLSSTCLRTLHSNAFNHLRILIELDLSQNELTQLSPDLFKATERLRVLILNDNPLTTLVAEQFPPLRYLKKLEISRCKLRSVSSFAFTNLRALESVNIQQNYLTHLHQSTFNLPLMNTLSLSDNPWYCDCGLREFHDWFVSSNLGNEEVLCLGPESKANISWSALKREDFFCSPSVLTSPTVIRTEVGADISFGCFVRGDPKPVVTWSLNRADVFNNIENGEITIHRYDIDNFNEYTDYYLNKSSQWVNISISNITNDLAGEWKCSAKSSVGDASTYLTILLPKARTARTATARVVPDYSAFFLIAGSMLAMTGVGFVSAYICWKTRRRRIPPSRSFTDQEKKLLDSSLAASCDRASGDMSLSYGFEMLDRSMSLDSEDNRCLEPVQITVEGPPGSFPPPPAEFALPAPYGNIFISVQVSGHNDEYPDLLGGGTTLPRRNTTCFLKSTYDNMGPRITAAGSSTWSLPGANDKNTVKENLLTTPLSNFSTEFTAL
Summary
Uniprot
H9JII6
A0A2W1BFD8
A0A2A4K9T7
A0A0L7LFL0
A0A194RJN9
A0A194Q8V2
+ More
A0A0L7LA24 A0A194Q786 A0A0N0PB35 A0A2A4K9Z8 A0A2W1BLB4 A0A2H1VLJ7 A0A212F4S8 D1ZZM3 A0A1W4XGW3 A0A2R7WTA1 A0A0T6B3T3 A0A2J7PG78 U4UPJ5 N6TG30 N6TSK3 T1ID69 A0A067QTN0 A0A1S3D2D1 E0VGJ7 J9K4B7 A0A2H8TNW6 A0A2S2PTV6 A0A087ZUM4 A0A0C9Q5G2 A0A154P804 A0A2A3EUI0 A0A2S2R7J8 A0A310SE10 A0A0L7R398 A0A0M9A4K9 E2BYJ8 E2AYH5 F4X3R4 A0A158P037 A0A2P8ZPB5 A0A1J1IQ56 A0A151XJA2 A0A195F2V7 A0A0J7P084 A0A084VX68 A0A026WRW2 A0A1B0D6N8 W8BGG8 A0A182IN95 A0A182GJN4 A0A336M660 A0A336LE96 A0A182U583 B0XEQ6 A0A2C9GPR6 Q7QFF0 A0A182UXE7 A0A182XDU1 A0A182Q5P7 A0A0L0C883 A0A182MWI9 A0A182K5Q4 A0A182W5H6 A0A182RMX3 A0A182XXT8 A0A182SGP7 W5JRT2 Q17L59 A0A1A9YQB6 A0A1B0BLI9 A0A1B0A1F8 A0A1B0FI23 K7ISF4 A0A182FJ78 A0A1I8MGN2 A0A1A9W199 A0A1A9ULQ3 A0A182PBJ1 A0A1I8Q759 A0A0K8V7H5 A0A195E055 A0A182N194 A0A195BGP3 B3P521 B4PNF9 B4QSP5 A0A0B4KHL5 A0A195CQZ4 B4IJ14 Q9V9V6 Q297R4 B4G2X2
A0A0L7LA24 A0A194Q786 A0A0N0PB35 A0A2A4K9Z8 A0A2W1BLB4 A0A2H1VLJ7 A0A212F4S8 D1ZZM3 A0A1W4XGW3 A0A2R7WTA1 A0A0T6B3T3 A0A2J7PG78 U4UPJ5 N6TG30 N6TSK3 T1ID69 A0A067QTN0 A0A1S3D2D1 E0VGJ7 J9K4B7 A0A2H8TNW6 A0A2S2PTV6 A0A087ZUM4 A0A0C9Q5G2 A0A154P804 A0A2A3EUI0 A0A2S2R7J8 A0A310SE10 A0A0L7R398 A0A0M9A4K9 E2BYJ8 E2AYH5 F4X3R4 A0A158P037 A0A2P8ZPB5 A0A1J1IQ56 A0A151XJA2 A0A195F2V7 A0A0J7P084 A0A084VX68 A0A026WRW2 A0A1B0D6N8 W8BGG8 A0A182IN95 A0A182GJN4 A0A336M660 A0A336LE96 A0A182U583 B0XEQ6 A0A2C9GPR6 Q7QFF0 A0A182UXE7 A0A182XDU1 A0A182Q5P7 A0A0L0C883 A0A182MWI9 A0A182K5Q4 A0A182W5H6 A0A182RMX3 A0A182XXT8 A0A182SGP7 W5JRT2 Q17L59 A0A1A9YQB6 A0A1B0BLI9 A0A1B0A1F8 A0A1B0FI23 K7ISF4 A0A182FJ78 A0A1I8MGN2 A0A1A9W199 A0A1A9ULQ3 A0A182PBJ1 A0A1I8Q759 A0A0K8V7H5 A0A195E055 A0A182N194 A0A195BGP3 B3P521 B4PNF9 B4QSP5 A0A0B4KHL5 A0A195CQZ4 B4IJ14 Q9V9V6 Q297R4 B4G2X2
Pubmed
19121390
28756777
26227816
26354079
22118469
18362917
+ More
19820115 23537049 24845553 20566863 20798317 21719571 21347285 29403074 24438588 24508170 30249741 24495485 26483478 12364791 14747013 17210077 26108605 25244985 20920257 23761445 17510324 20075255 25315136 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085
19820115 23537049 24845553 20566863 20798317 21719571 21347285 29403074 24438588 24508170 30249741 24495485 26483478 12364791 14747013 17210077 26108605 25244985 20920257 23761445 17510324 20075255 25315136 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085
EMBL
BABH01027129
KZ150185
PZC72465.1
NWSH01000018
PCG80735.1
PCG80736.1
+ More
JTDY01001333 KOB74165.1 KQ460297 KPJ16161.1 KQ459337 KPJ01395.1 JTDY01002017 KOB72317.1 KPJ01393.1 KQ461056 KPJ09928.1 PCG80734.1 PZC72463.1 ODYU01003222 SOQ41688.1 AGBW02010318 OWR48741.1 KQ971338 EFA02390.1 KK855489 PTY22793.1 LJIG01016129 KRT81527.1 NEVH01025635 PNF15333.1 KB632308 ERL91945.1 APGK01029141 KB740725 ENN79349.1 APGK01002864 KB734997 ENN83469.1 ACPB03002840 KK853033 KDR12250.1 DS235147 EEB12503.1 ABLF02041079 GFXV01003369 GFXV01003806 GFXV01007561 MBW15174.1 MBW15611.1 MBW19366.1 GGMR01020196 MBY32815.1 GBYB01009278 JAG79045.1 KQ434839 KZC08055.1 KZ288185 PBC34866.1 GGMS01016735 MBY85938.1 KQ768090 OAD53216.1 KQ414663 KOC65309.1 KQ435758 KOX75823.1 GL451483 EFN79232.1 GL443910 EFN61510.1 GL888624 EGI58861.1 ADTU01005290 PYGN01000004 PSN58346.1 CVRI01000057 CRL02373.1 KQ982074 KYQ60492.1 KQ981856 KYN34512.1 LBMM01000566 KMQ98050.1 ATLV01017851 KE525195 KFB42562.1 KK107139 QOIP01000003 EZA57844.1 RLU24780.1 AJVK01012101 GAMC01008713 JAB97842.1 JXUM01068626 KQ562510 KXJ75749.1 UFQT01000589 SSX25520.1 UFQS01004090 UFQT01004090 SSX16168.1 SSX35494.1 DS232855 EDS26165.1 APCN01001309 AAAB01008846 EAA06629.5 AXCN02001237 JRES01000755 KNC28648.1 AXCM01009331 ADMH02000610 ETN65635.1 CH477218 EAT47436.1 JXJN01016400 CCAG010018193 GDHF01017749 GDHF01011331 JAI34565.1 JAI40983.1 KQ979955 KYN18481.1 KQ976490 KYM83344.1 CH954182 EDV52935.1 CM000160 EDW99241.2 CM000364 EDX15142.1 AE014297 AGB96529.1 KQ977394 KYN03065.1 CH480846 EDW50967.1 BT100194 AAF57176.1 ACY40028.1 CM000070 EAL28141.4 CH479179 EDW24167.1
JTDY01001333 KOB74165.1 KQ460297 KPJ16161.1 KQ459337 KPJ01395.1 JTDY01002017 KOB72317.1 KPJ01393.1 KQ461056 KPJ09928.1 PCG80734.1 PZC72463.1 ODYU01003222 SOQ41688.1 AGBW02010318 OWR48741.1 KQ971338 EFA02390.1 KK855489 PTY22793.1 LJIG01016129 KRT81527.1 NEVH01025635 PNF15333.1 KB632308 ERL91945.1 APGK01029141 KB740725 ENN79349.1 APGK01002864 KB734997 ENN83469.1 ACPB03002840 KK853033 KDR12250.1 DS235147 EEB12503.1 ABLF02041079 GFXV01003369 GFXV01003806 GFXV01007561 MBW15174.1 MBW15611.1 MBW19366.1 GGMR01020196 MBY32815.1 GBYB01009278 JAG79045.1 KQ434839 KZC08055.1 KZ288185 PBC34866.1 GGMS01016735 MBY85938.1 KQ768090 OAD53216.1 KQ414663 KOC65309.1 KQ435758 KOX75823.1 GL451483 EFN79232.1 GL443910 EFN61510.1 GL888624 EGI58861.1 ADTU01005290 PYGN01000004 PSN58346.1 CVRI01000057 CRL02373.1 KQ982074 KYQ60492.1 KQ981856 KYN34512.1 LBMM01000566 KMQ98050.1 ATLV01017851 KE525195 KFB42562.1 KK107139 QOIP01000003 EZA57844.1 RLU24780.1 AJVK01012101 GAMC01008713 JAB97842.1 JXUM01068626 KQ562510 KXJ75749.1 UFQT01000589 SSX25520.1 UFQS01004090 UFQT01004090 SSX16168.1 SSX35494.1 DS232855 EDS26165.1 APCN01001309 AAAB01008846 EAA06629.5 AXCN02001237 JRES01000755 KNC28648.1 AXCM01009331 ADMH02000610 ETN65635.1 CH477218 EAT47436.1 JXJN01016400 CCAG010018193 GDHF01017749 GDHF01011331 JAI34565.1 JAI40983.1 KQ979955 KYN18481.1 KQ976490 KYM83344.1 CH954182 EDV52935.1 CM000160 EDW99241.2 CM000364 EDX15142.1 AE014297 AGB96529.1 KQ977394 KYN03065.1 CH480846 EDW50967.1 BT100194 AAF57176.1 ACY40028.1 CM000070 EAL28141.4 CH479179 EDW24167.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053240
UP000053268
UP000007151
+ More
UP000007266 UP000192223 UP000235965 UP000030742 UP000019118 UP000015103 UP000027135 UP000079169 UP000009046 UP000007819 UP000005203 UP000076502 UP000242457 UP000053825 UP000053105 UP000008237 UP000000311 UP000007755 UP000005205 UP000245037 UP000183832 UP000075809 UP000078541 UP000036403 UP000030765 UP000053097 UP000279307 UP000092462 UP000075880 UP000069940 UP000249989 UP000075902 UP000002320 UP000075840 UP000007062 UP000075903 UP000076407 UP000075886 UP000037069 UP000075883 UP000075881 UP000075920 UP000075900 UP000076408 UP000075901 UP000000673 UP000008820 UP000092443 UP000092460 UP000092445 UP000092444 UP000002358 UP000069272 UP000095301 UP000091820 UP000078200 UP000075885 UP000095300 UP000078492 UP000075884 UP000078540 UP000008711 UP000002282 UP000000304 UP000000803 UP000078542 UP000001292 UP000001819 UP000008744
UP000007266 UP000192223 UP000235965 UP000030742 UP000019118 UP000015103 UP000027135 UP000079169 UP000009046 UP000007819 UP000005203 UP000076502 UP000242457 UP000053825 UP000053105 UP000008237 UP000000311 UP000007755 UP000005205 UP000245037 UP000183832 UP000075809 UP000078541 UP000036403 UP000030765 UP000053097 UP000279307 UP000092462 UP000075880 UP000069940 UP000249989 UP000075902 UP000002320 UP000075840 UP000007062 UP000075903 UP000076407 UP000075886 UP000037069 UP000075883 UP000075881 UP000075920 UP000075900 UP000076408 UP000075901 UP000000673 UP000008820 UP000092443 UP000092460 UP000092445 UP000092444 UP000002358 UP000069272 UP000095301 UP000091820 UP000078200 UP000075885 UP000095300 UP000078492 UP000075884 UP000078540 UP000008711 UP000002282 UP000000304 UP000000803 UP000078542 UP000001292 UP000001819 UP000008744
Interpro
IPR013783
Ig-like_fold
+ More
IPR000483 Cys-rich_flank_reg_C
IPR007110 Ig-like_dom
IPR013098 Ig_I-set
IPR032675 LRR_dom_sf
IPR003591 Leu-rich_rpt_typical-subtyp
IPR001611 Leu-rich_rpt
IPR036179 Ig-like_dom_sf
IPR003599 Ig_sub
IPR003598 Ig_sub2
IPR009100 AcylCoA_DH/oxidase_NM_dom
IPR006091 Acyl-CoA_Oxase/DH_cen-dom
IPR009075 AcylCo_DH/oxidase_C
IPR036250 AcylCo_DH-like_C
IPR037069 AcylCoA_DH/ox_N_sf
IPR000483 Cys-rich_flank_reg_C
IPR007110 Ig-like_dom
IPR013098 Ig_I-set
IPR032675 LRR_dom_sf
IPR003591 Leu-rich_rpt_typical-subtyp
IPR001611 Leu-rich_rpt
IPR036179 Ig-like_dom_sf
IPR003599 Ig_sub
IPR003598 Ig_sub2
IPR009100 AcylCoA_DH/oxidase_NM_dom
IPR006091 Acyl-CoA_Oxase/DH_cen-dom
IPR009075 AcylCo_DH/oxidase_C
IPR036250 AcylCo_DH-like_C
IPR037069 AcylCoA_DH/ox_N_sf
Gene 3D
ProteinModelPortal
H9JII6
A0A2W1BFD8
A0A2A4K9T7
A0A0L7LFL0
A0A194RJN9
A0A194Q8V2
+ More
A0A0L7LA24 A0A194Q786 A0A0N0PB35 A0A2A4K9Z8 A0A2W1BLB4 A0A2H1VLJ7 A0A212F4S8 D1ZZM3 A0A1W4XGW3 A0A2R7WTA1 A0A0T6B3T3 A0A2J7PG78 U4UPJ5 N6TG30 N6TSK3 T1ID69 A0A067QTN0 A0A1S3D2D1 E0VGJ7 J9K4B7 A0A2H8TNW6 A0A2S2PTV6 A0A087ZUM4 A0A0C9Q5G2 A0A154P804 A0A2A3EUI0 A0A2S2R7J8 A0A310SE10 A0A0L7R398 A0A0M9A4K9 E2BYJ8 E2AYH5 F4X3R4 A0A158P037 A0A2P8ZPB5 A0A1J1IQ56 A0A151XJA2 A0A195F2V7 A0A0J7P084 A0A084VX68 A0A026WRW2 A0A1B0D6N8 W8BGG8 A0A182IN95 A0A182GJN4 A0A336M660 A0A336LE96 A0A182U583 B0XEQ6 A0A2C9GPR6 Q7QFF0 A0A182UXE7 A0A182XDU1 A0A182Q5P7 A0A0L0C883 A0A182MWI9 A0A182K5Q4 A0A182W5H6 A0A182RMX3 A0A182XXT8 A0A182SGP7 W5JRT2 Q17L59 A0A1A9YQB6 A0A1B0BLI9 A0A1B0A1F8 A0A1B0FI23 K7ISF4 A0A182FJ78 A0A1I8MGN2 A0A1A9W199 A0A1A9ULQ3 A0A182PBJ1 A0A1I8Q759 A0A0K8V7H5 A0A195E055 A0A182N194 A0A195BGP3 B3P521 B4PNF9 B4QSP5 A0A0B4KHL5 A0A195CQZ4 B4IJ14 Q9V9V6 Q297R4 B4G2X2
A0A0L7LA24 A0A194Q786 A0A0N0PB35 A0A2A4K9Z8 A0A2W1BLB4 A0A2H1VLJ7 A0A212F4S8 D1ZZM3 A0A1W4XGW3 A0A2R7WTA1 A0A0T6B3T3 A0A2J7PG78 U4UPJ5 N6TG30 N6TSK3 T1ID69 A0A067QTN0 A0A1S3D2D1 E0VGJ7 J9K4B7 A0A2H8TNW6 A0A2S2PTV6 A0A087ZUM4 A0A0C9Q5G2 A0A154P804 A0A2A3EUI0 A0A2S2R7J8 A0A310SE10 A0A0L7R398 A0A0M9A4K9 E2BYJ8 E2AYH5 F4X3R4 A0A158P037 A0A2P8ZPB5 A0A1J1IQ56 A0A151XJA2 A0A195F2V7 A0A0J7P084 A0A084VX68 A0A026WRW2 A0A1B0D6N8 W8BGG8 A0A182IN95 A0A182GJN4 A0A336M660 A0A336LE96 A0A182U583 B0XEQ6 A0A2C9GPR6 Q7QFF0 A0A182UXE7 A0A182XDU1 A0A182Q5P7 A0A0L0C883 A0A182MWI9 A0A182K5Q4 A0A182W5H6 A0A182RMX3 A0A182XXT8 A0A182SGP7 W5JRT2 Q17L59 A0A1A9YQB6 A0A1B0BLI9 A0A1B0A1F8 A0A1B0FI23 K7ISF4 A0A182FJ78 A0A1I8MGN2 A0A1A9W199 A0A1A9ULQ3 A0A182PBJ1 A0A1I8Q759 A0A0K8V7H5 A0A195E055 A0A182N194 A0A195BGP3 B3P521 B4PNF9 B4QSP5 A0A0B4KHL5 A0A195CQZ4 B4IJ14 Q9V9V6 Q297R4 B4G2X2
PDB
6F2O
E-value=1.77711e-16,
Score=212
Ontologies
GO
Topology
SignalP
Position: 1 - 21,
Likelihood: 0.998657
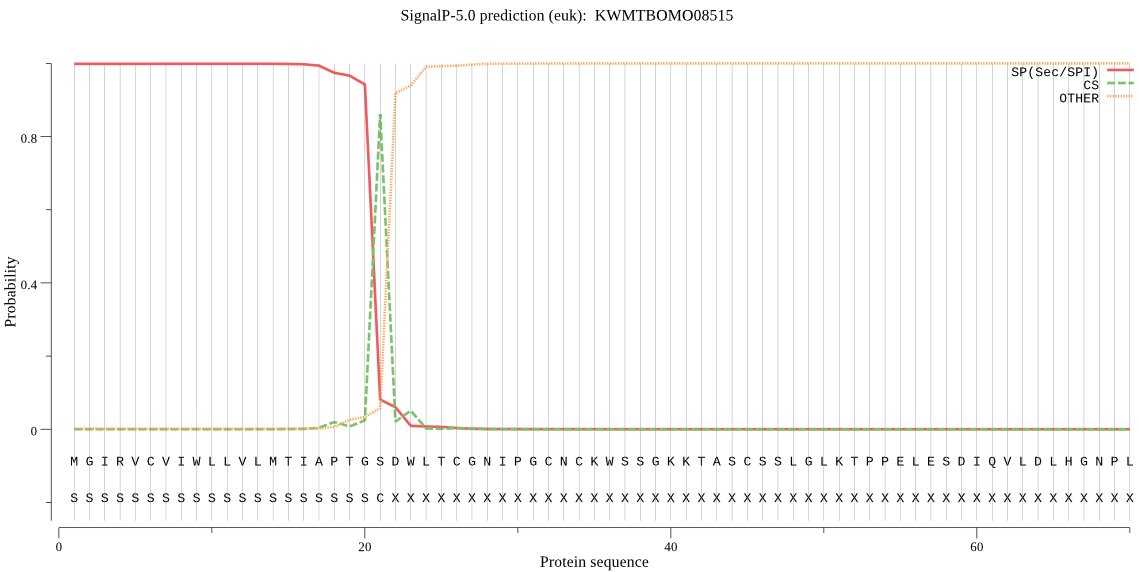
Length:
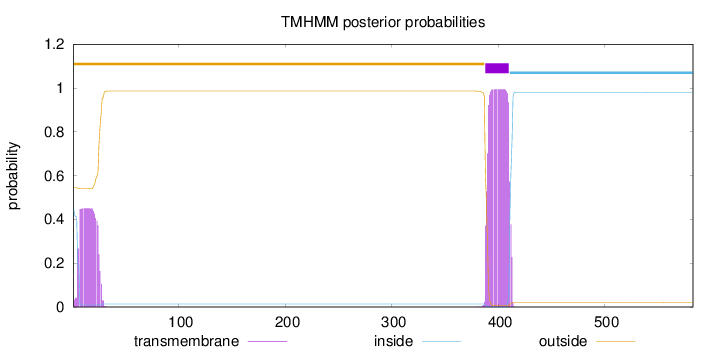
583
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
31.6422099999999
Exp number, first 60 AAs:
9.10939
Total prob of N-in:
0.45421
outside
1 - 387
TMhelix
388 - 410
inside
411 - 583
Population Genetic Test Statistics
Pi
280.496717
Theta
188.91171
Tajima's D
1.423441
CLR
0.230218
CSRT
0.767311634418279
Interpretation
Uncertain
