Gene
KWMTBOMO08507
Pre Gene Modal
BGIBMGA009340
Annotation
PREDICTED:_speckle-type_POZ_protein-like_[Plutella_xylostella]
Full name
Speckle-type POZ protein-like
Alternative Name
HIB homolog 2
Location in the cell
Cytoplasmic Reliability : 1.314 Nuclear Reliability : 1.768
Sequence
CDS
ATGTCGAGCACAAATTATACAACGCGCACGGAGGGACAGACAAAAATATGTACTATAATATGGACCATACCTACGTTCGTCAATCTGTTGGAAAATGTGACCAAGAGGGAATTTCGAGTTGAAAAAGCTAAAGACCCAAAAACGGAATTTACTCACTCTAAGTTCCAATTGAAAATGCAATTTTTGGGCCGTTATAATGACATAATAGAGATTTCTTATCTATCACCAAATCCGGTTTTTTTGAAGTCCATTCTAACAATTTGTTTAAAAAGATTTGAAGAAAGAACAATAGTAATAAAGGAGTATCACACGGTTCAGGCAAACAAATGGCAGTACTTGGCTACATTATTCAAACGAGATATAGCAAGCTATGATGGCCGTGATATATTCCTCCTTAGTGATGGAAGTTTACGACTTAAAGTACAATTTATGGTCACCAATGATATAAAAATAGACCGTTCAACAGTACCTGAAGCACAACTCAGCAATGACTTAGAGAATTTACTCAATGATGGGTTGTTCTCTGATGTAATAATCAAGTCTGCAGAGGGTATCGAGTTCAAAGTCCACAAGGCTGTGTTGGCCAGCCGTAGTGCTGTTCTAAAAGCTCATTTTGAACACAACACCACAGAATGCTACACTAATGTTATTGAATCGCTTTTAGAATCAACAGTATTACAAGAAGTACTAACATTTATTTATAGTGACAAAGCTCCTAGAGTTGATGACATTCCAGAAAAGCTTCTAGCAGCTGCCGATTACTACCAACTTAACAGACTAAAGAGCTTATGCGAGGAAGCATTGCATAAAAAATTGACAGTAGAGAATGCTATAGAAACATTACAATTAGCTGATTTACATTCAGCAAATACATTGAAACAACTCACTCTTGAATTTATAAAAGATGGACAAGCAAAGCTTATAACAAAAACTGAAGGATGGGCCCAAGTAAAATCTGTTGATTTAATCAAAAGAATCTATGAATACATCATGGCAGATGATTTCGATGCTGATATAAAATGA
Protein
MSSTNYTTRTEGQTKICTIIWTIPTFVNLLENVTKREFRVEKAKDPKTEFTHSKFQLKMQFLGRYNDIIEISYLSPNPVFLKSILTICLKRFEERTIVIKEYHTVQANKWQYLATLFKRDIASYDGRDIFLLSDGSLRLKVQFMVTNDIKIDRSTVPEAQLSNDLENLLNDGLFSDVIIKSAEGIEFKVHKAVLASRSAVLKAHFEHNTTECYTNVIESLLESTVLQEVLTFIYSDKAPRVDDIPEKLLAAADYYQLNRLKSLCEEALHKKLTVENAIETLQLADLHSANTLKQLTLEFIKDGQAKLITKTEGWAQVKSVDLIKRIYEYIMADDFDADIK
Summary
Description
Component of a cullin-RING-based BCR (BTB-CUL3-RBX1) E3 ubiquitin-protein ligase complex that mediates the ubiquitination and subsequent proteasomal degradation of target proteins, but with relatively low efficiency. Cullin-RING-based BCR (BTB-CUL3-RBX1) E3 ubiquitin-protein ligase complexes containing homodimeric SPOPL or the heterodimer formed by SPOP and SPOPL are less efficient than ubiquitin ligase complexes containing only SPOP. May function to down-regulate the activity of cullin-RING-based BCR (BTB-CUL3-RBX1) E3 ubiquitin-protein ligase complexes that contain SPOP (By similarity).
Subunit
Homodimer. Heterodimer with SPOP. Component of cullin-RING-based BCR (BTB-CUL3-RBX1) E3 ubiquitin-protein ligase complexes containing homodimeric SPOPL or the heterodimer formed by SPOP and SPOPL. Interacts with CUL3 and H2AFY (By similarity).
Similarity
Belongs to the Tdpoz family.
Keywords
Alternative splicing
Complete proteome
Nucleus
Reference proteome
Ubl conjugation pathway
Feature
chain Speckle-type POZ protein-like
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9JIJ0
A0A1E1VZH7
A0A2H1V014
A0A2W1B8X2
A0A2A4JXA8
A0A194Q7N3
+ More
A0A0N0PEL5 S4P812 A0A0L7LKY6 A0A0L7L2A6 A0A0L7L4E2 A0A0L7LJ77 A0A0L7KZP8 A0A0L7L4M6 A0A0L7KJH1 A0A0L7K445 A0A0L7KZG9 A0A0L7LBZ1 A0A0L7KML2 A0A0L7LKA0 A0A0L7L5K4 A0A0L7K1Z2 A0A0L7KDP9 A0A0L7KZA4 A0A0L7KZ63 A0A0L7LA12 A0A0L7K4F4 A0A0L7KKQ4 A0A0L7LUK8 A0A0L7LE50 A0A0L7LAC9 V4ABW3 A0A194RS66 B2RZC7 Q2M2N2 A0A194PFB6 A0A1U8CJW3 G3IE83 H9JSI4 A0A1E1VZR2 A0A2P6K791 R7UX04 A0A1E1WJF8 A0A1E1W5S0 A0A2K5QG11 A0A212F9Z7 A0A267FS63 A0A437BNH0
A0A0N0PEL5 S4P812 A0A0L7LKY6 A0A0L7L2A6 A0A0L7L4E2 A0A0L7LJ77 A0A0L7KZP8 A0A0L7L4M6 A0A0L7KJH1 A0A0L7K445 A0A0L7KZG9 A0A0L7LBZ1 A0A0L7KML2 A0A0L7LKA0 A0A0L7L5K4 A0A0L7K1Z2 A0A0L7KDP9 A0A0L7KZA4 A0A0L7KZ63 A0A0L7LA12 A0A0L7K4F4 A0A0L7KKQ4 A0A0L7LUK8 A0A0L7LE50 A0A0L7LAC9 V4ABW3 A0A194RS66 B2RZC7 Q2M2N2 A0A194PFB6 A0A1U8CJW3 G3IE83 H9JSI4 A0A1E1VZR2 A0A2P6K791 R7UX04 A0A1E1WJF8 A0A1E1W5S0 A0A2K5QG11 A0A212F9Z7 A0A267FS63 A0A437BNH0
Pubmed
EMBL
BABH01027092
GDQN01010970
JAT80084.1
ODYU01000049
SOQ34159.1
KZ150432
+ More
PZC70895.1 NWSH01000476 PCG76133.1 KQ459337 KPJ01399.1 KQ459928 KPJ19303.1 GAIX01006156 JAA86404.1 JTDY01000719 KOB76102.1 JTDY01003440 KOB69555.1 JTDY01003036 KOB70280.1 JTDY01000901 KOB75502.1 JTDY01004130 KOB68551.1 KOB70281.1 JTDY01009633 KOB63191.1 JTDY01012845 KOB52227.1 KOB68550.1 JTDY01001850 KOB72721.1 JTDY01008517 KOB64537.1 JTDY01000759 KOB75988.1 JTDY01002755 KOB70783.1 JTDY01017745 KOB51836.1 JTDY01010117 KOB61024.1 KOB68552.1 KOB68553.1 JTDY01002058 KOB72215.1 JTDY01011163 KOB57450.1 JTDY01009464 KOB63459.1 JTDY01000095 KOB78896.1 JTDY01001520 KOB73665.1 JTDY01001956 KOB72453.1 KB201847 ESO94307.1 KQ459984 KPJ18911.1 AABR07051193 BC167106 CH474001 AAI67106.1 EDL93676.1 AK162056 AL773534 CH466542 BC111867 KQ459606 KPI91389.1 JH002147 RAZU01000235 EGW06666.1 RLQ63825.1 BABH01027901 BABH01027902 GDQN01011347 GDQN01010836 JAT79707.1 JAT80218.1 MWRG01022350 PRD22211.1 AMQN01005851 KB296957 ELU11118.1 GDQN01003932 JAT87122.1 GDQN01008738 JAT82316.1 AGBW02009539 OWR50554.1 NIVC01002410 NIVC01000807 PAA58105.1 PAA76635.1 RSAL01000027 RVE51918.1
PZC70895.1 NWSH01000476 PCG76133.1 KQ459337 KPJ01399.1 KQ459928 KPJ19303.1 GAIX01006156 JAA86404.1 JTDY01000719 KOB76102.1 JTDY01003440 KOB69555.1 JTDY01003036 KOB70280.1 JTDY01000901 KOB75502.1 JTDY01004130 KOB68551.1 KOB70281.1 JTDY01009633 KOB63191.1 JTDY01012845 KOB52227.1 KOB68550.1 JTDY01001850 KOB72721.1 JTDY01008517 KOB64537.1 JTDY01000759 KOB75988.1 JTDY01002755 KOB70783.1 JTDY01017745 KOB51836.1 JTDY01010117 KOB61024.1 KOB68552.1 KOB68553.1 JTDY01002058 KOB72215.1 JTDY01011163 KOB57450.1 JTDY01009464 KOB63459.1 JTDY01000095 KOB78896.1 JTDY01001520 KOB73665.1 JTDY01001956 KOB72453.1 KB201847 ESO94307.1 KQ459984 KPJ18911.1 AABR07051193 BC167106 CH474001 AAI67106.1 EDL93676.1 AK162056 AL773534 CH466542 BC111867 KQ459606 KPI91389.1 JH002147 RAZU01000235 EGW06666.1 RLQ63825.1 BABH01027901 BABH01027902 GDQN01011347 GDQN01010836 JAT79707.1 JAT80218.1 MWRG01022350 PRD22211.1 AMQN01005851 KB296957 ELU11118.1 GDQN01003932 JAT87122.1 GDQN01008738 JAT82316.1 AGBW02009539 OWR50554.1 NIVC01002410 NIVC01000807 PAA58105.1 PAA76635.1 RSAL01000027 RVE51918.1
Proteomes
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JIJ0
A0A1E1VZH7
A0A2H1V014
A0A2W1B8X2
A0A2A4JXA8
A0A194Q7N3
+ More
A0A0N0PEL5 S4P812 A0A0L7LKY6 A0A0L7L2A6 A0A0L7L4E2 A0A0L7LJ77 A0A0L7KZP8 A0A0L7L4M6 A0A0L7KJH1 A0A0L7K445 A0A0L7KZG9 A0A0L7LBZ1 A0A0L7KML2 A0A0L7LKA0 A0A0L7L5K4 A0A0L7K1Z2 A0A0L7KDP9 A0A0L7KZA4 A0A0L7KZ63 A0A0L7LA12 A0A0L7K4F4 A0A0L7KKQ4 A0A0L7LUK8 A0A0L7LE50 A0A0L7LAC9 V4ABW3 A0A194RS66 B2RZC7 Q2M2N2 A0A194PFB6 A0A1U8CJW3 G3IE83 H9JSI4 A0A1E1VZR2 A0A2P6K791 R7UX04 A0A1E1WJF8 A0A1E1W5S0 A0A2K5QG11 A0A212F9Z7 A0A267FS63 A0A437BNH0
A0A0N0PEL5 S4P812 A0A0L7LKY6 A0A0L7L2A6 A0A0L7L4E2 A0A0L7LJ77 A0A0L7KZP8 A0A0L7L4M6 A0A0L7KJH1 A0A0L7K445 A0A0L7KZG9 A0A0L7LBZ1 A0A0L7KML2 A0A0L7LKA0 A0A0L7L5K4 A0A0L7K1Z2 A0A0L7KDP9 A0A0L7KZA4 A0A0L7KZ63 A0A0L7LA12 A0A0L7K4F4 A0A0L7KKQ4 A0A0L7LUK8 A0A0L7LE50 A0A0L7LAC9 V4ABW3 A0A194RS66 B2RZC7 Q2M2N2 A0A194PFB6 A0A1U8CJW3 G3IE83 H9JSI4 A0A1E1VZR2 A0A2P6K791 R7UX04 A0A1E1WJF8 A0A1E1W5S0 A0A2K5QG11 A0A212F9Z7 A0A267FS63 A0A437BNH0
PDB
4J8Z
E-value=6.25887e-31,
Score=334
Ontologies
GO
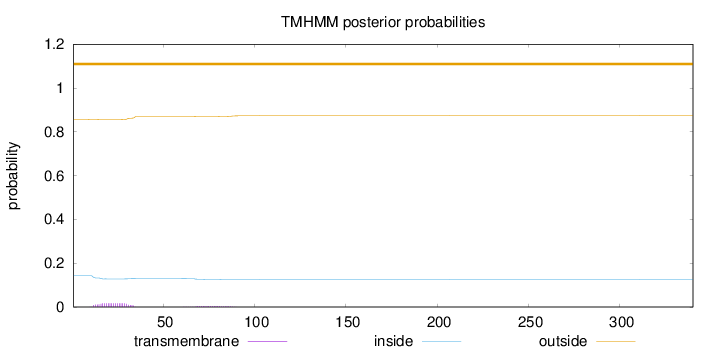
Topology
Subcellular location
Nucleus
Length:
340
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.402410000000001
Exp number, first 60 AAs:
0.31925
Total prob of N-in:
0.14270
outside
1 - 340
Population Genetic Test Statistics
Pi
188.522167
Theta
190.96841
Tajima's D
-0.083092
CLR
348.629733
CSRT
0.348782560871956
Interpretation
Uncertain
