Gene
KWMTBOMO08503 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009359
Annotation
AP-1_complex_subunit_mu-1_[Papilio_xuthus]
Location in the cell
Golgi Reliability : 3.264
Sequence
CDS
ATGTCTTCTTCGGCTATATACATACTAGATGTCAAAGGTAAAGTTCTCATTTCGAGAAATTACCGTGGTGATGTTGATCTGGGTGTGATCGACAAATTCATGCCTTTACTGATGGAAAAGGAAGAAGAGGGAATGTTAACGCCTTTATTACAGACTAGTGAATGTACATTCGCCTATATCAAAACCAACAACTTGTACATTGTATCAACCACAAAGAAAAATGCTAATATTGCACTTGTGTTTGTATTCTTGTACAAAATTGTTGAAGTTATGACTGAGTACTTCAAAGAATTAGAGGAAGAAAGCATTCGAGACAACTTTGTGGTTATCTACGAGCTACTTGACGAACTCATAGACTTTGGATACCCTCAGACCACTGATAGCAAAATTCTACAAGAATACATCACCCAAGAAGGTCATAAACTTGAAATGCAGCCCAAAATACCCATGGCAGTTACCAATGCTGTGTCGTGGAGGTCCGAAGGCATAAAATATAGAAAGAATGAAGTCTTCTTAGATGTTATCGAGTCTGTAAATTTATTAGCCAACTCAAATGGTAATGTACTGAGGAGTGAGATTGTTGGAGCTATTAAAATGAGAGTGTACTTGTCTGGTATGCCAGAATTACGTTTAGGCTTAAACGATAAGGTCTTATTTGAAAGTACCGGCAGAGGAAAATCAAAGTCTGTGGAGTTAGAAGATGTGAAGTTCCATCAATGTGTTAGATTATCACGTTTTGAAAATGATAGGACCATCTCATTTATACCCCCTGATGGTGAATTTGAATTGATGTCATACCGACTCAACACTCATGTAAAACCTCTGATTTGGATAGAGTCTGTTATTGAGCGTCATGCTCATTCAAGAGTTGAATACATGATAAAGGCTAAGTCTCAGTTTAAGAGACGATCAACTGCGAATAACGTTGAAATCATTATACCCGTTCCTGCTGATGCTGATTCACCAAAATTCAAGACAACAATTGGTAGCGTTAAATATACACCAGAACAAAATGCGATCACATGGTCAATCAAATCATTTCCAGGAGGCAAGGAGTACTTAATGAGAGCTCATTTTGGTCTGCCTTCTGTTGAATGTGAGGAAGTTGATGGAAAGCCCCCAATTCAAGTAAAATTTGAAATACCGTATTTCACAACTTCTGGAATTCAAGTAAGATATCTGAAAATTATTGAAAAAAGTGGATACCAAGCCCTACCTTGGGTAAGGTACATTACTCAGAATGGTGATTATCAGTTAAGAACAAATTAA
Protein
MSSSAIYILDVKGKVLISRNYRGDVDLGVIDKFMPLLMEKEEEGMLTPLLQTSECTFAYIKTNNLYIVSTTKKNANIALVFVFLYKIVEVMTEYFKELEEESIRDNFVVIYELLDELIDFGYPQTTDSKILQEYITQEGHKLEMQPKIPMAVTNAVSWRSEGIKYRKNEVFLDVIESVNLLANSNGNVLRSEIVGAIKMRVYLSGMPELRLGLNDKVLFESTGRGKSKSVELEDVKFHQCVRLSRFENDRTISFIPPDGEFELMSYRLNTHVKPLIWIESVIERHAHSRVEYMIKAKSQFKRRSTANNVEIIIPVPADADSPKFKTTIGSVKYTPEQNAITWSIKSFPGGKEYLMRAHFGLPSVECEEVDGKPPIQVKFEIPYFTTSGIQVRYLKIIEKSGYQALPWVRYITQNGDYQLRTN
Summary
Similarity
Belongs to the adaptor complexes medium subunit family.
Uniprot
A0A194Q782
A0A2W1B235
A0A2A4JWH7
A0A0N1ICB4
S4PE66
A0A212EKY2
+ More
I4DJC5 A0A0L7L7E3 A0A1B6LT94 A0A1B6FF13 A0A0T6BG22 H9JIK9 A0A1B6CJD8 A0A1W4X626 A0A1Y1NI13 A0A2J7QMU4 A0A2J7QMY1 D7EIL0 A0A0K8TTJ0 A0A0P4VZC3 R4G4C8 A0A2R7X799 A0A224XD59 A0A023FAR0 A0A069DTP8 J3JW23 A0A0A9YQD1 T1E8Q4 A0A1Q3FCQ2 A0A2M4AET0 A0A182MCA1 A0A182PTP9 A0A182QP26 A0A182V538 A0A182N388 W5JJB2 A0A182KG05 A0A182U493 A0A182W1Q8 B0XEY9 A0A084WNT9 A0A182J4I7 A0A182WXD8 A0A182LQ92 Q7Q2W7 A0A182FBQ7 A0A182RVE7 A0A182HY14 Q16S47 A0A023ERX9 U5EMT5 E0VCL1 A0A1B0CLT9 A0A1S3DRR3 A0A1L8DVQ0 A0A1B0D9R8 A0A2M4BPR5 Q173K2 K7IUN8 A0A232F2G8 A0A0L7QYW5 A0A2A3E7B3 V9IJ79 A0A087ZV20 E9J5F4 A0A1J1IL99 A0A151IBD3 A0A154P3V9 A0A0J7KNX8 E2BMK0 E2AN72 A0A195BSA0 A0A158NV92 A0A026WQL8 A0A151J1E6 A0A195FX11 A0A0L0C9L2 A0A1L8ECW1 A0A1I8PXG6 T1J6Q9 A0A3B0JP30 T1D419 B4K659 B4M4B2 Q295J6 B4GDS5 A0A336KHC5 B4NB34 T1PB43 A0A0M4ER13 A0A1A9XW96 A0A1A9VFL4 A0A1B0BL73 D3TMQ4 B4QWK7 B4PUQ5 B4HJY3 B3NZQ4 A0A1W4VMV4 B3M2F9
I4DJC5 A0A0L7L7E3 A0A1B6LT94 A0A1B6FF13 A0A0T6BG22 H9JIK9 A0A1B6CJD8 A0A1W4X626 A0A1Y1NI13 A0A2J7QMU4 A0A2J7QMY1 D7EIL0 A0A0K8TTJ0 A0A0P4VZC3 R4G4C8 A0A2R7X799 A0A224XD59 A0A023FAR0 A0A069DTP8 J3JW23 A0A0A9YQD1 T1E8Q4 A0A1Q3FCQ2 A0A2M4AET0 A0A182MCA1 A0A182PTP9 A0A182QP26 A0A182V538 A0A182N388 W5JJB2 A0A182KG05 A0A182U493 A0A182W1Q8 B0XEY9 A0A084WNT9 A0A182J4I7 A0A182WXD8 A0A182LQ92 Q7Q2W7 A0A182FBQ7 A0A182RVE7 A0A182HY14 Q16S47 A0A023ERX9 U5EMT5 E0VCL1 A0A1B0CLT9 A0A1S3DRR3 A0A1L8DVQ0 A0A1B0D9R8 A0A2M4BPR5 Q173K2 K7IUN8 A0A232F2G8 A0A0L7QYW5 A0A2A3E7B3 V9IJ79 A0A087ZV20 E9J5F4 A0A1J1IL99 A0A151IBD3 A0A154P3V9 A0A0J7KNX8 E2BMK0 E2AN72 A0A195BSA0 A0A158NV92 A0A026WQL8 A0A151J1E6 A0A195FX11 A0A0L0C9L2 A0A1L8ECW1 A0A1I8PXG6 T1J6Q9 A0A3B0JP30 T1D419 B4K659 B4M4B2 Q295J6 B4GDS5 A0A336KHC5 B4NB34 T1PB43 A0A0M4ER13 A0A1A9XW96 A0A1A9VFL4 A0A1B0BL73 D3TMQ4 B4QWK7 B4PUQ5 B4HJY3 B3NZQ4 A0A1W4VMV4 B3M2F9
Pubmed
26354079
28756777
23622113
22118469
22651552
26227816
+ More
19121390 28004739 18362917 19820115 26369729 27129103 25474469 26334808 22516182 23537049 25401762 26823975 20920257 23761445 24438588 20966253 12364791 14747013 17210077 17510324 24945155 26483478 20566863 20075255 28648823 21282665 20798317 21347285 24508170 30249741 26108605 24330624 17994087 18057021 15632085 25315136 20353571 17550304
19121390 28004739 18362917 19820115 26369729 27129103 25474469 26334808 22516182 23537049 25401762 26823975 20920257 23761445 24438588 20966253 12364791 14747013 17210077 17510324 24945155 26483478 20566863 20075255 28648823 21282665 20798317 21347285 24508170 30249741 26108605 24330624 17994087 18057021 15632085 25315136 20353571 17550304
EMBL
KQ459337
KPJ01402.1
KZ150432
PZC70892.1
NWSH01000476
PCG76136.1
+ More
KQ459928 KPJ19300.1 GAIX01003291 JAA89269.1 AGBW02014173 OWR42142.1 AK401393 BAM18015.1 JTDY01002447 KOB71403.1 GEBQ01013110 JAT26867.1 GECZ01021075 JAS48694.1 LJIG01000650 KRT86290.1 BABH01027089 BABH01027090 BABH01027091 BABH01027092 GEDC01023692 JAS13606.1 GEZM01001852 JAV97491.1 NEVH01013202 PNF29914.1 PNF29913.1 KQ971307 EFA12022.1 GDAI01000348 JAI17255.1 GDKW01001706 JAI54889.1 ACPB03006269 GAHY01001480 JAA76030.1 KK858185 PTY27667.1 GFTR01006079 JAW10347.1 GBBI01000583 JAC18129.1 GBGD01001589 JAC87300.1 APGK01026024 BT127441 KB740612 KB632373 AEE62403.1 ENN79999.1 ERL93769.1 GBHO01009763 GBRD01003248 GDHC01008174 JAG33841.1 JAG62573.1 JAQ10455.1 GAMD01002568 JAA99022.1 GFDL01009753 JAV25292.1 GGFK01005936 MBW39257.1 AXCM01000696 AXCN02001122 ADMH02001015 GGFL01004206 ETN64452.1 MBW68384.1 DS232883 EDS26337.1 ATLV01024718 KE525359 KFB51883.1 AXCP01005858 AAAB01008966 EAA13067.4 APCN01004467 CH477683 EAT37288.1 JXUM01141942 GAPW01001633 KQ569335 JAC11965.1 KXJ68657.1 GANO01000876 JAB58995.1 DS235059 EEB11117.1 AJWK01017609 GFDF01003576 JAV10508.1 AJVK01028425 AJVK01028426 GGFJ01005770 MBW54911.1 CH477422 EAT41228.1 AAZX01002620 NNAY01001217 OXU24699.1 KQ414687 KOC63742.1 KZ288345 PBC27590.1 JR050102 AEY61178.1 GL768153 EFZ11942.1 CVRI01000054 CRK99854.1 KQ978106 KYM96960.1 KQ434804 KZC06024.1 LBMM01004968 KMQ91949.1 GL449234 EFN83071.1 GL441103 EFN65114.1 KQ976417 KYM89895.1 ADTU01027039 KK107128 QOIP01000006 EZA58320.1 RLU22016.1 KQ980575 KYN15482.1 KQ981193 KYN45190.1 JRES01000819 KNC28139.1 GFDG01002317 JAV16482.1 JH431883 OUUW01000008 SPP83885.1 GALA01001133 JAA93719.1 CH933806 EDW14109.1 KRG00866.1 CH940652 EDW59473.1 KRF78884.1 CM000070 EAL28715.1 CH479182 EDW34586.1 UFQS01000376 UFQT01000376 SSX03342.1 SSX23708.1 CH964232 EDW80998.1 KA645952 AFP60581.1 CP012526 ALC46872.1 JXJN01016236 CCAG010001933 EZ422706 ADD18982.1 CM000364 EDX13611.1 CM000160 EDW96672.1 CH480815 EDW42867.1 CH954181 EDV49764.1 CH902617 EDV43412.1 KPU80266.1
KQ459928 KPJ19300.1 GAIX01003291 JAA89269.1 AGBW02014173 OWR42142.1 AK401393 BAM18015.1 JTDY01002447 KOB71403.1 GEBQ01013110 JAT26867.1 GECZ01021075 JAS48694.1 LJIG01000650 KRT86290.1 BABH01027089 BABH01027090 BABH01027091 BABH01027092 GEDC01023692 JAS13606.1 GEZM01001852 JAV97491.1 NEVH01013202 PNF29914.1 PNF29913.1 KQ971307 EFA12022.1 GDAI01000348 JAI17255.1 GDKW01001706 JAI54889.1 ACPB03006269 GAHY01001480 JAA76030.1 KK858185 PTY27667.1 GFTR01006079 JAW10347.1 GBBI01000583 JAC18129.1 GBGD01001589 JAC87300.1 APGK01026024 BT127441 KB740612 KB632373 AEE62403.1 ENN79999.1 ERL93769.1 GBHO01009763 GBRD01003248 GDHC01008174 JAG33841.1 JAG62573.1 JAQ10455.1 GAMD01002568 JAA99022.1 GFDL01009753 JAV25292.1 GGFK01005936 MBW39257.1 AXCM01000696 AXCN02001122 ADMH02001015 GGFL01004206 ETN64452.1 MBW68384.1 DS232883 EDS26337.1 ATLV01024718 KE525359 KFB51883.1 AXCP01005858 AAAB01008966 EAA13067.4 APCN01004467 CH477683 EAT37288.1 JXUM01141942 GAPW01001633 KQ569335 JAC11965.1 KXJ68657.1 GANO01000876 JAB58995.1 DS235059 EEB11117.1 AJWK01017609 GFDF01003576 JAV10508.1 AJVK01028425 AJVK01028426 GGFJ01005770 MBW54911.1 CH477422 EAT41228.1 AAZX01002620 NNAY01001217 OXU24699.1 KQ414687 KOC63742.1 KZ288345 PBC27590.1 JR050102 AEY61178.1 GL768153 EFZ11942.1 CVRI01000054 CRK99854.1 KQ978106 KYM96960.1 KQ434804 KZC06024.1 LBMM01004968 KMQ91949.1 GL449234 EFN83071.1 GL441103 EFN65114.1 KQ976417 KYM89895.1 ADTU01027039 KK107128 QOIP01000006 EZA58320.1 RLU22016.1 KQ980575 KYN15482.1 KQ981193 KYN45190.1 JRES01000819 KNC28139.1 GFDG01002317 JAV16482.1 JH431883 OUUW01000008 SPP83885.1 GALA01001133 JAA93719.1 CH933806 EDW14109.1 KRG00866.1 CH940652 EDW59473.1 KRF78884.1 CM000070 EAL28715.1 CH479182 EDW34586.1 UFQS01000376 UFQT01000376 SSX03342.1 SSX23708.1 CH964232 EDW80998.1 KA645952 AFP60581.1 CP012526 ALC46872.1 JXJN01016236 CCAG010001933 EZ422706 ADD18982.1 CM000364 EDX13611.1 CM000160 EDW96672.1 CH480815 EDW42867.1 CH954181 EDV49764.1 CH902617 EDV43412.1 KPU80266.1
Proteomes
UP000053268
UP000218220
UP000053240
UP000007151
UP000037510
UP000005204
+ More
UP000192223 UP000235965 UP000007266 UP000015103 UP000019118 UP000030742 UP000075883 UP000075885 UP000075886 UP000075903 UP000075884 UP000000673 UP000075881 UP000075902 UP000075920 UP000002320 UP000030765 UP000075880 UP000076407 UP000075882 UP000007062 UP000069272 UP000075900 UP000075840 UP000008820 UP000069940 UP000249989 UP000009046 UP000092461 UP000079169 UP000092462 UP000002358 UP000215335 UP000053825 UP000242457 UP000005203 UP000183832 UP000078542 UP000076502 UP000036403 UP000008237 UP000000311 UP000078540 UP000005205 UP000053097 UP000279307 UP000078492 UP000078541 UP000037069 UP000095300 UP000268350 UP000009192 UP000008792 UP000001819 UP000008744 UP000007798 UP000095301 UP000092553 UP000092443 UP000078200 UP000092460 UP000092444 UP000000304 UP000002282 UP000001292 UP000008711 UP000192221 UP000007801
UP000192223 UP000235965 UP000007266 UP000015103 UP000019118 UP000030742 UP000075883 UP000075885 UP000075886 UP000075903 UP000075884 UP000000673 UP000075881 UP000075902 UP000075920 UP000002320 UP000030765 UP000075880 UP000076407 UP000075882 UP000007062 UP000069272 UP000075900 UP000075840 UP000008820 UP000069940 UP000249989 UP000009046 UP000092461 UP000079169 UP000092462 UP000002358 UP000215335 UP000053825 UP000242457 UP000005203 UP000183832 UP000078542 UP000076502 UP000036403 UP000008237 UP000000311 UP000078540 UP000005205 UP000053097 UP000279307 UP000078492 UP000078541 UP000037069 UP000095300 UP000268350 UP000009192 UP000008792 UP000001819 UP000008744 UP000007798 UP000095301 UP000092553 UP000092443 UP000078200 UP000092460 UP000092444 UP000000304 UP000002282 UP000001292 UP000008711 UP000192221 UP000007801
PRIDE
Interpro
ProteinModelPortal
A0A194Q782
A0A2W1B235
A0A2A4JWH7
A0A0N1ICB4
S4PE66
A0A212EKY2
+ More
I4DJC5 A0A0L7L7E3 A0A1B6LT94 A0A1B6FF13 A0A0T6BG22 H9JIK9 A0A1B6CJD8 A0A1W4X626 A0A1Y1NI13 A0A2J7QMU4 A0A2J7QMY1 D7EIL0 A0A0K8TTJ0 A0A0P4VZC3 R4G4C8 A0A2R7X799 A0A224XD59 A0A023FAR0 A0A069DTP8 J3JW23 A0A0A9YQD1 T1E8Q4 A0A1Q3FCQ2 A0A2M4AET0 A0A182MCA1 A0A182PTP9 A0A182QP26 A0A182V538 A0A182N388 W5JJB2 A0A182KG05 A0A182U493 A0A182W1Q8 B0XEY9 A0A084WNT9 A0A182J4I7 A0A182WXD8 A0A182LQ92 Q7Q2W7 A0A182FBQ7 A0A182RVE7 A0A182HY14 Q16S47 A0A023ERX9 U5EMT5 E0VCL1 A0A1B0CLT9 A0A1S3DRR3 A0A1L8DVQ0 A0A1B0D9R8 A0A2M4BPR5 Q173K2 K7IUN8 A0A232F2G8 A0A0L7QYW5 A0A2A3E7B3 V9IJ79 A0A087ZV20 E9J5F4 A0A1J1IL99 A0A151IBD3 A0A154P3V9 A0A0J7KNX8 E2BMK0 E2AN72 A0A195BSA0 A0A158NV92 A0A026WQL8 A0A151J1E6 A0A195FX11 A0A0L0C9L2 A0A1L8ECW1 A0A1I8PXG6 T1J6Q9 A0A3B0JP30 T1D419 B4K659 B4M4B2 Q295J6 B4GDS5 A0A336KHC5 B4NB34 T1PB43 A0A0M4ER13 A0A1A9XW96 A0A1A9VFL4 A0A1B0BL73 D3TMQ4 B4QWK7 B4PUQ5 B4HJY3 B3NZQ4 A0A1W4VMV4 B3M2F9
I4DJC5 A0A0L7L7E3 A0A1B6LT94 A0A1B6FF13 A0A0T6BG22 H9JIK9 A0A1B6CJD8 A0A1W4X626 A0A1Y1NI13 A0A2J7QMU4 A0A2J7QMY1 D7EIL0 A0A0K8TTJ0 A0A0P4VZC3 R4G4C8 A0A2R7X799 A0A224XD59 A0A023FAR0 A0A069DTP8 J3JW23 A0A0A9YQD1 T1E8Q4 A0A1Q3FCQ2 A0A2M4AET0 A0A182MCA1 A0A182PTP9 A0A182QP26 A0A182V538 A0A182N388 W5JJB2 A0A182KG05 A0A182U493 A0A182W1Q8 B0XEY9 A0A084WNT9 A0A182J4I7 A0A182WXD8 A0A182LQ92 Q7Q2W7 A0A182FBQ7 A0A182RVE7 A0A182HY14 Q16S47 A0A023ERX9 U5EMT5 E0VCL1 A0A1B0CLT9 A0A1S3DRR3 A0A1L8DVQ0 A0A1B0D9R8 A0A2M4BPR5 Q173K2 K7IUN8 A0A232F2G8 A0A0L7QYW5 A0A2A3E7B3 V9IJ79 A0A087ZV20 E9J5F4 A0A1J1IL99 A0A151IBD3 A0A154P3V9 A0A0J7KNX8 E2BMK0 E2AN72 A0A195BSA0 A0A158NV92 A0A026WQL8 A0A151J1E6 A0A195FX11 A0A0L0C9L2 A0A1L8ECW1 A0A1I8PXG6 T1J6Q9 A0A3B0JP30 T1D419 B4K659 B4M4B2 Q295J6 B4GDS5 A0A336KHC5 B4NB34 T1PB43 A0A0M4ER13 A0A1A9XW96 A0A1A9VFL4 A0A1B0BL73 D3TMQ4 B4QWK7 B4PUQ5 B4HJY3 B3NZQ4 A0A1W4VMV4 B3M2F9
PDB
6DFF
E-value=0,
Score=1871
Ontologies
GO
PANTHER
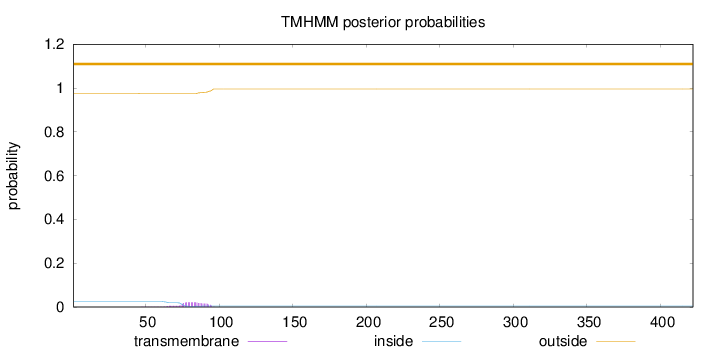
Topology
Length:
422
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.429770000000001
Exp number, first 60 AAs:
0.00256
Total prob of N-in:
0.02448
outside
1 - 422
Population Genetic Test Statistics
Pi
215.324213
Theta
196.079965
Tajima's D
0.221661
CLR
1.918511
CSRT
0.43682815859207
Interpretation
Uncertain
