Gene
KWMTBOMO08502
Pre Gene Modal
BGIBMGA009341
Annotation
PREDICTED:_peptidyl-prolyl_cis-trans_isomerase_D_isoform_X1_[Bombyx_mori]
Full name
Peptidyl-prolyl cis-trans isomerase D
Alternative Name
40 kDa peptidyl-prolyl cis-trans isomerase
Cyclophilin-40
Rotamase D
Cyclophilin-related protein
Estrogen receptor-binding cyclophilin
Cyclophilin-40
Rotamase D
Cyclophilin-related protein
Estrogen receptor-binding cyclophilin
Location in the cell
Cytoplasmic Reliability : 1.645 Nuclear Reliability : 1.865
Sequence
CDS
ATGAGTTCCGACAGGCGATCACAAACGAATCCTTTTGTTTATTTGGACATCTCTATTGATGGAGTATCAGCTGGACGAATCGTGATCGAACTGAGGAGTGACGTAGTCCCGAAAACCGCAGAGAACTTCCGGGCGCTTTGTACGGGCGAGAAGGGCATTGGAGTTTACGGGAAACCCTTACACTACAAAGGAGTCAGATTCCATAAAGCGGTTAATCAGTTCATGGTGCAGGGCGGAGACATAGTCAACGGCGACGGCAGCGGCGGCGAGAGCATATACGGCCCGAAATTCGAAGACGAGAATTTTACTCTTACGGCAAAGTCTGGGGTACTGGCGATGGCGAACGAAGGTCGACCGGACACAAACAGTTCGCAGTTCTGTATAACGACGATACCGTGCCCGCACATAAACGGCACTAATGTTATATTCGGGGAGGTGGTCGCCGGATTCGAACTGGTCGACGAGATGCAGAGATACGGCGAAGGGGACGACGGGAGGCCCGGGGCGGAATGCGTGATCGAAGACTGCGGCGAAATAACAGAGCAGAACTGGGACGTGTGCTGCAGGGATGGCACCTCCGACAAAATACCCGAATTCCCGACGGACTGGAGGAATATTGGAGCCGTTTCGTTAGACGATTTAATATCGTGCATAAGGGACGTCAAGGCAGCCGGCAATGGAATGTTCAGCGAGCGTCGGTACAAAGCCGCGATCAGAAAGTACCGCAAGTGCCTCAGATACGTCGACTACGCTTTGAGTATAGTACAAGGAATGCACGAGAAAGAGACAGAACATATAATAGATTTAACATCAACGTTTATACTGCAGTGTAATTTGAATCTGGCCGCGTGTTACTCGAGAACCGAAGACTACAGAGCGTGTATCGAGTGCTGTACGCAGGTGCTGGACAGAAGCCCTCGCAACGAGAAGGCCCTGTACCGGCGGGGCCAAGCGAACTTCGCCCTTAAGAACTACGAGGCCGCGCTGTCCGATCTGAGGCGGGCCGAGCGCGCGTCGCCGCACAATCGCGCCGTGCTCGCGCTGCTGGACGAGGTGCGCCGCTGCAGCCGCCGCTACAACGAGCTGCAGAAACAGAGGCTCTCAAAGTTTTTTCGTGAGCAAAAAGAGCGCGCCGCCGCCGCCAACAACTGA
Protein
MSSDRRSQTNPFVYLDISIDGVSAGRIVIELRSDVVPKTAENFRALCTGEKGIGVYGKPLHYKGVRFHKAVNQFMVQGGDIVNGDGSGGESIYGPKFEDENFTLTAKSGVLAMANEGRPDTNSSQFCITTIPCPHINGTNVIFGEVVAGFELVDEMQRYGEGDDGRPGAECVIEDCGEITEQNWDVCCRDGTSDKIPEFPTDWRNIGAVSLDDLISCIRDVKAAGNGMFSERRYKAAIRKYRKCLRYVDYALSIVQGMHEKETEHIIDLTSTFILQCNLNLAACYSRTEDYRACIECCTQVLDRSPRNEKALYRRGQANFALKNYEAALSDLRRAERASPHNRAVLALLDEVRRCSRRYNELQKQRLSKFFREQKERAAAANN
Summary
Description
PPIase that catalyzes the cis-trans isomerization of proline imidic peptide bonds in oligopeptides and may therefore assist protein folding. Proposed to act as a co-chaperone in HSP90 complexes such as in unligated steroid receptors heterocomplexes. Different co-chaperones seem to compete for association with HSP90 thus establishing distinct HSP90-co-chaperone-receptor complexes with the potential to exert tissue-specific receptor activity control. May have a preference for estrogen receptor complexes and is not found in glucocorticoid receptor complexes. May be involved in cytoplasmic dynein-dependent movement of the receptor from the cytoplasm to the nucleus. May regulate MYB by inhibiting its DNA-binding activity. Involved in regulation of AHR signaling by promoting the formation of the AHR:ARNT dimer; the function is independent of HSP90 but requires the chaperone activity region. Involved in regulation of UV radiation-induced apoptosis.
PPIase that catalyzes the cis-trans isomerization of proline imidic peptide bonds in oligopeptides and may therefore assist protein folding. Proposed to act as a co-chaperone in HSP90 complexes such as in unligated steroid receptors heterocomplexes. Different co-chaperones seem to compete for association with HSP90 thus establishing distinct HSP90-co-chaperone-receptor complexes with the potential to exert tissue-specific receptor activity control. May have a preference for estrogen receptor complexes and is not found in glucocorticoid receptor complexes. May be involved in cytoplasmic dynein-dependent movement of the receptor from the cytoplasm to the nucleus. May regulate MYB by inhibiting its DNA-binding activity. Involved in regulation of AHR signaling by promoting the formation of the AHR:ARNT dimer; the function is independent of HSP90 but requires the chaperone activity. Involved in regulation of UV radiation-induced apoptosis.
PPIase that catalyzes the cis-trans isomerization of proline imidic peptide bonds in oligopeptides and may therefore assist protein folding (PubMed:11350175, PubMed:20676357). Proposed to act as a co-chaperone in HSP90 complexes such as in unligated steroid receptors heterocomplexes. Different co-chaperones seem to compete for association with HSP90 thus establishing distinct HSP90-co-chaperone-receptor complexes with the potential to exert tissue-specific receptor activity control. May have a preference for estrogen receptor complexes and is not found in glucocorticoid receptor complexes. May be involved in cytoplasmic dynein-dependent movement of the receptor from the cytoplasm to the nucleus. May regulate MYB by inhibiting its DNA-binding activity. Involved in regulation of AHR signaling by promoting the formation of the AHR:ARNT dimer; the function is independent of HSP90 but requires the chaperone activity. Involved in regulation of UV radiation-induced apoptosis. Promotes cell viability in anaplastic lymphoma kinase-positive anaplastic large-cell lymphoma (ALK+ ALCL) cell lines.
(Microbial infection) May be involved in hepatitis C virus (HCV) replication and release.
PPIase that catalyzes the cis-trans isomerization of proline imidic peptide bonds in oligopeptides and may therefore assist protein folding. Proposed to act as a co-chaperone in HSP90 complexes such as in unligated steroid receptors heterocomplexes. Different co-chaperones seem to compete for association with HSP90 thus establishing distinct HSP90-co-chaperone-receptor complexes with the potential to exert tissue-specific receptor activity control. May have a preference for estrogen receptor complexes and is not found in glucocorticoid receptor complexes. May be involved in cytoplasmic dynein-dependent movement of the receptor from the cytoplasm to the nucleus. May regulate MYB by inhibiting its DNA-binding activity. Involved in regulation of AHR signaling by promoting the formation of the AHR:ARNT dimer; the function is independent of HSP90 but requires the chaperone activity. Involved in regulation of UV radiation-induced apoptosis.
PPIase that catalyzes the cis-trans isomerization of proline imidic peptide bonds in oligopeptides and may therefore assist protein folding (PubMed:11350175, PubMed:20676357). Proposed to act as a co-chaperone in HSP90 complexes such as in unligated steroid receptors heterocomplexes. Different co-chaperones seem to compete for association with HSP90 thus establishing distinct HSP90-co-chaperone-receptor complexes with the potential to exert tissue-specific receptor activity control. May have a preference for estrogen receptor complexes and is not found in glucocorticoid receptor complexes. May be involved in cytoplasmic dynein-dependent movement of the receptor from the cytoplasm to the nucleus. May regulate MYB by inhibiting its DNA-binding activity. Involved in regulation of AHR signaling by promoting the formation of the AHR:ARNT dimer; the function is independent of HSP90 but requires the chaperone activity. Involved in regulation of UV radiation-induced apoptosis. Promotes cell viability in anaplastic lymphoma kinase-positive anaplastic large-cell lymphoma (ALK+ ALCL) cell lines.
(Microbial infection) May be involved in hepatitis C virus (HCV) replication and release.
Catalytic Activity
[protein]-peptidylproline (omega=180) = [protein]-peptidylproline (omega=0)
Subunit
Identified in ESR1 or NR3C1/GCR steroid receptor-chaperone complexes. Found in HSP90 chaperone complexes with kinase clients LCK or EIF2AK1. Two monomers associate with one HSP90 homodimer. Interacts with HSP90AA1. Interacts with HSP90AB1; PPID and FKBP4 compete for binding to HSP90AB1 and the interaction is mutually exclusive with the PPID:HSPA8 interaction. Interacts with HSPA8; PPID and STIP1 but not FKBP4 compete for binding to HSPA8 and the interaction is mutually exclusive with the PPID:HSP90AB1 interaction. Interacts with S100A1 and S100A2; the interactions dissociate the PPID:HSP90AA1 interaction. Interacts with S100A6. Interacts with MYB, ILF2, XRCC6, RACK1 and RPS3. Interacts with cytoplasmic dynein 1 intermediate chain (DYNC1I1 or DYNC1I2).
Identified in ESR1 or NR3C1/GCR steroid receptor-chaperone complexes. Found in HSP90 chaperone complexes with kinase clients LCK or EIF2AK1. Two monomers associate with one HSP90 homodimer. Interacts with HSP90AA1. Interacts with HSP90AB1; PPID and FKBP4 compete for binding to HSP90AB1 and the interaction is mutually exclusive with the PPID:HSPA8 interaction. Interacts with HSPA8; PPID and STIP1 but not FKBP4 compete for binding to HSPA8 and the interaction is mutually exclusive with the PPID:HSP90AB1 interaction. Interacts with S100A1 and S100A2; the interactions dissociate the PPID:HSP90AA1 interaction. Interacts with S100A6. Interacts with MYB, ILF2, XRCC6, RACK1 and RPS3. Interacts with cytoplasmic dynein 1 intermediate chain (DYNC1I1 or DYNC1I2) (By similarity).
Identified in ESR1 or NR3C1/GCR steroid receptor-chaperone complexes. Found in HSP90 chaperone complexes with kinase clients LCK or EIF2AK1. Two monomers associate with one HSP90 homodimer. Interacts with HSP90AA1. Interacts with HSP90AB1; PPID and FKBP4 compete for binding to HSP90AB1 and the interaction is mutually exclusive with the PPID:HSPA8 interaction. Interacts with HSPA8; PPID and STIP1 but not FKBP4 compete for binding to HSPA8 and the interaction is mutually exclusive with the PPID:HSP90AB1 interaction. Interacts with S100A1 and S100A2; the interactions dissociate the PPID:HSP90AA1 interaction. Interacts with S100A6. Interacts with MYB, ILF2, XRCC6, RACK1 and RPS3. Interacts with cytoplasmic dynein 1 intermediate chain (DYNC1I1 or DYNC1I2) (By similarity).
Similarity
Belongs to the cyclophilin-type PPIase family. PPIase D subfamily.
Keywords
Acetylation
Apoptosis
Chaperone
Complete proteome
Cytoplasm
Direct protein sequencing
Isomerase
Nucleus
Phosphoprotein
Protein transport
Reference proteome
Repeat
Rotamase
TPR repeat
Transport
3D-structure
Polymorphism
Feature
chain Peptidyl-prolyl cis-trans isomerase D
sequence variant In dbSNP:rs2070631.
sequence variant In dbSNP:rs2070631.
Uniprot
H9JIJ1
A0A2H1VXD7
A0A2A4JVR6
A0A2W1B6N7
A0A212EKZ2
A0A1I8N2U0
+ More
A0A1A9WTN3 A0A1I8PTX9 B4QN98 B4HL81 K7J6J5 B3NCJ9 A0A084VCY4 Q9VT21 A0A067RL00 A0A0M3QW77 A0A1W4UMC2 A0A182RQ92 W8BMC2 D3TPY2 A0A0K8W0V4 A0A1A9UFZ0 A0A0L0C8M1 B4PEE8 A0A182Q0W1 Q641F7 A0A250XZF9 A0A034W0S0 D2A2T9 A0A182N574 H2PEM6 A0A088AV72 A0A2K6SY51 A0A0A9Z9H3 A0A2K6QXE7 A0A0K8T5L8 U5EYW6 A0A2Y9DVV3 A0A2K5CX43 A0A2K5SDA6 A0A2K5U0U8 A0A2K6A6Y4 A0A2K5MFH5 H9EVU2 A0A096N788 F7IL08 G1KSN5 A0A1S3GAL8 K7GBA2 G7NSZ6 A0A1U7QL23 G1R3M8 A0A2A3ETC7 Q3UB60 A0A3L7IHP4 G7P6H6 I7GL27 Q9CR16 A0A1U7SYN1 A0A2K5I5F7 A0A2K5UHD9 A0A182J317 A0A3Q2US37 A0A2K6KDJ4 I3IY94 A0A182MNW8 H0UVB7 G3TDA2 L5MLJ1 A0A1Z5L541 Q6P894 A0A1L8HUD3 A0A0M8ZTM2 A0A452J5W3 B4LHR5 Q6DGG0 A0A2J7PGH4 K9K4R4 S7MJ67 A0A2R9A916 H2QQC9 B4IY92 A0A2Y9F907 A0A2K6DSW2 A0A340WDL9 A0A1A7XW11 G3RAF0 A0A2Y9NSC0 A0A2K6QXE6 P26882 E5KN55 Q08752 A0A3P9NPY5 L8HPW4 F7DFM9 F1RTY6 A0A182HJD1 Q6FGM6 E5KN59 A0A341BJF3
A0A1A9WTN3 A0A1I8PTX9 B4QN98 B4HL81 K7J6J5 B3NCJ9 A0A084VCY4 Q9VT21 A0A067RL00 A0A0M3QW77 A0A1W4UMC2 A0A182RQ92 W8BMC2 D3TPY2 A0A0K8W0V4 A0A1A9UFZ0 A0A0L0C8M1 B4PEE8 A0A182Q0W1 Q641F7 A0A250XZF9 A0A034W0S0 D2A2T9 A0A182N574 H2PEM6 A0A088AV72 A0A2K6SY51 A0A0A9Z9H3 A0A2K6QXE7 A0A0K8T5L8 U5EYW6 A0A2Y9DVV3 A0A2K5CX43 A0A2K5SDA6 A0A2K5U0U8 A0A2K6A6Y4 A0A2K5MFH5 H9EVU2 A0A096N788 F7IL08 G1KSN5 A0A1S3GAL8 K7GBA2 G7NSZ6 A0A1U7QL23 G1R3M8 A0A2A3ETC7 Q3UB60 A0A3L7IHP4 G7P6H6 I7GL27 Q9CR16 A0A1U7SYN1 A0A2K5I5F7 A0A2K5UHD9 A0A182J317 A0A3Q2US37 A0A2K6KDJ4 I3IY94 A0A182MNW8 H0UVB7 G3TDA2 L5MLJ1 A0A1Z5L541 Q6P894 A0A1L8HUD3 A0A0M8ZTM2 A0A452J5W3 B4LHR5 Q6DGG0 A0A2J7PGH4 K9K4R4 S7MJ67 A0A2R9A916 H2QQC9 B4IY92 A0A2Y9F907 A0A2K6DSW2 A0A340WDL9 A0A1A7XW11 G3RAF0 A0A2Y9NSC0 A0A2K6QXE6 P26882 E5KN55 Q08752 A0A3P9NPY5 L8HPW4 F7DFM9 F1RTY6 A0A182HJD1 Q6FGM6 E5KN59 A0A341BJF3
EC Number
5.2.1.8
Pubmed
19121390
28756777
22118469
25315136
17994087
22936249
+ More
20075255 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24845553 24495485 20353571 26108605 17550304 28087693 25348373 18362917 19820115 25401762 25362486 25319552 25243066 17381049 22002653 10349636 11042159 11076861 11217851 12466851 16141073 29704459 17194215 16141072 15489334 9195923 18771283 21183079 23806337 25186727 21993624 28528879 20431018 27762356 28562605 22722832 16136131 22398555 8514757 8509368 1544925 8621687 9915798 15497503 16650407 20188096 11377203 20843780 8812478 14702039 12665801 9659917 11525244 11350175 12145316 14580201 18708059 18669648 19690332 20676357 19932913 21146485 21269460 21711559 22681779 23220213 23186163 24275569 22751099 19892987
20075255 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24845553 24495485 20353571 26108605 17550304 28087693 25348373 18362917 19820115 25401762 25362486 25319552 25243066 17381049 22002653 10349636 11042159 11076861 11217851 12466851 16141073 29704459 17194215 16141072 15489334 9195923 18771283 21183079 23806337 25186727 21993624 28528879 20431018 27762356 28562605 22722832 16136131 22398555 8514757 8509368 1544925 8621687 9915798 15497503 16650407 20188096 11377203 20843780 8812478 14702039 12665801 9659917 11525244 11350175 12145316 14580201 18708059 18669648 19690332 20676357 19932913 21146485 21269460 21711559 22681779 23220213 23186163 24275569 22751099 19892987
EMBL
BABH01027085
BABH01027086
ODYU01004703
SOQ44864.1
NWSH01000476
PCG76137.1
+ More
KZ150432 PZC70891.1 AGBW02014173 OWR42141.1 CM000363 CM002912 EDX09881.1 KMY98694.1 CH480815 EDW40900.1 AAZX01001318 CH954178 EDV51229.1 ATLV01011003 KE524632 KFB35828.1 AE014296 AY058303 AAF50235.1 AAL13532.1 KK852614 KDR20206.1 CP012525 ALC43663.1 GAMC01004310 JAC02246.1 EZ423484 ADD19760.1 GDHF01007546 JAI44768.1 JRES01000850 KNC27744.1 CM000159 EDW92986.1 AXCN02000166 BC082380 AAH82380.1 GFFV01003187 JAV36758.1 GAKP01011574 JAC47378.1 KQ971339 EFA01462.1 ABGA01009582 ABGA01009583 NDHI03003405 PNJ63182.1 GBHO01002540 JAG41064.1 GBRD01005003 JAG60818.1 GANO01001773 JAB58098.1 AQIA01056827 JU322744 JU322745 JU473029 JU473030 JU473031 JV045027 AFE66501.1 AFH29833.1 AFI35098.1 AHZZ02028075 GAMT01007734 GAMS01010887 GAMR01005540 GAMQ01001654 GAMQ01001653 JAB04127.1 JAB12249.1 JAB28392.1 JAB40197.1 AGCU01077551 AGCU01077552 AGCU01077553 AGCU01077554 AGCU01077555 AGCU01077556 CM001273 EHH30637.1 ADFV01178721 KZ288192 PBC34301.1 AK150520 AK151091 BAE30104.1 RAZU01000055 RLQ77121.1 CM001280 EHH54044.1 AB172112 BAE89174.1 AK003402 AK013919 AK051597 BC011499 BC019778 AQIA01084131 AERX01009909 AXCM01009363 AAKN02012071 KB098555 ELK38663.1 GFJQ02004839 JAW02131.1 AAMC01036942 BC061335 AAH61335.1 CM004466 OCT99660.1 KQ435850 KOX70900.1 CH940647 EDW69618.2 BC076386 NEVH01025635 PNF15420.1 JL616521 AEH58728.1 KE161484 EPQ04151.1 AJFE02078546 AACZ04005089 GABC01004090 GABD01006515 GABE01011524 GABE01011523 NBAG03000243 JAA07248.1 JAA26585.1 JAA33215.1 PNI63369.1 CH916366 EDV96542.1 HADW01020609 HADX01006315 SBP22009.1 CABD030034465 D14074 BC113318 L11668 HQ205146 HQ205147 HQ205148 HQ205149 HQ205151 HQ205152 HQ205153 HQ205154 HQ205155 HQ205156 HQ205157 HQ205158 HQ205159 HQ205160 HQ205161 HQ205162 HQ205163 HQ205165 HQ205166 HQ205168 HQ205169 HQ205170 HQ205171 HQ205172 HQ205173 HQ205174 HQ205175 HQ205176 HQ205177 HQ205178 HQ205179 HQ205180 HQ205181 HQ205182 HQ205183 HQ205184 HQ205185 HM005460 ADP90614.1 AEE61058.1 L11667 D63861 AY714221 AK313929 CH471056 BC030707 JH883514 ELR46335.1 AEMK02000065 APCN01002147 CR542081 CAG46878.1 HQ205150 HQ205164 HQ205167 ADP90618.1
KZ150432 PZC70891.1 AGBW02014173 OWR42141.1 CM000363 CM002912 EDX09881.1 KMY98694.1 CH480815 EDW40900.1 AAZX01001318 CH954178 EDV51229.1 ATLV01011003 KE524632 KFB35828.1 AE014296 AY058303 AAF50235.1 AAL13532.1 KK852614 KDR20206.1 CP012525 ALC43663.1 GAMC01004310 JAC02246.1 EZ423484 ADD19760.1 GDHF01007546 JAI44768.1 JRES01000850 KNC27744.1 CM000159 EDW92986.1 AXCN02000166 BC082380 AAH82380.1 GFFV01003187 JAV36758.1 GAKP01011574 JAC47378.1 KQ971339 EFA01462.1 ABGA01009582 ABGA01009583 NDHI03003405 PNJ63182.1 GBHO01002540 JAG41064.1 GBRD01005003 JAG60818.1 GANO01001773 JAB58098.1 AQIA01056827 JU322744 JU322745 JU473029 JU473030 JU473031 JV045027 AFE66501.1 AFH29833.1 AFI35098.1 AHZZ02028075 GAMT01007734 GAMS01010887 GAMR01005540 GAMQ01001654 GAMQ01001653 JAB04127.1 JAB12249.1 JAB28392.1 JAB40197.1 AGCU01077551 AGCU01077552 AGCU01077553 AGCU01077554 AGCU01077555 AGCU01077556 CM001273 EHH30637.1 ADFV01178721 KZ288192 PBC34301.1 AK150520 AK151091 BAE30104.1 RAZU01000055 RLQ77121.1 CM001280 EHH54044.1 AB172112 BAE89174.1 AK003402 AK013919 AK051597 BC011499 BC019778 AQIA01084131 AERX01009909 AXCM01009363 AAKN02012071 KB098555 ELK38663.1 GFJQ02004839 JAW02131.1 AAMC01036942 BC061335 AAH61335.1 CM004466 OCT99660.1 KQ435850 KOX70900.1 CH940647 EDW69618.2 BC076386 NEVH01025635 PNF15420.1 JL616521 AEH58728.1 KE161484 EPQ04151.1 AJFE02078546 AACZ04005089 GABC01004090 GABD01006515 GABE01011524 GABE01011523 NBAG03000243 JAA07248.1 JAA26585.1 JAA33215.1 PNI63369.1 CH916366 EDV96542.1 HADW01020609 HADX01006315 SBP22009.1 CABD030034465 D14074 BC113318 L11668 HQ205146 HQ205147 HQ205148 HQ205149 HQ205151 HQ205152 HQ205153 HQ205154 HQ205155 HQ205156 HQ205157 HQ205158 HQ205159 HQ205160 HQ205161 HQ205162 HQ205163 HQ205165 HQ205166 HQ205168 HQ205169 HQ205170 HQ205171 HQ205172 HQ205173 HQ205174 HQ205175 HQ205176 HQ205177 HQ205178 HQ205179 HQ205180 HQ205181 HQ205182 HQ205183 HQ205184 HQ205185 HM005460 ADP90614.1 AEE61058.1 L11667 D63861 AY714221 AK313929 CH471056 BC030707 JH883514 ELR46335.1 AEMK02000065 APCN01002147 CR542081 CAG46878.1 HQ205150 HQ205164 HQ205167 ADP90618.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000095301
UP000091820
UP000095300
+ More
UP000000304 UP000001292 UP000002358 UP000008711 UP000030765 UP000000803 UP000027135 UP000092553 UP000192221 UP000075900 UP000078200 UP000037069 UP000002282 UP000075886 UP000007266 UP000075884 UP000001595 UP000005203 UP000233220 UP000233200 UP000248480 UP000233020 UP000233040 UP000233100 UP000233140 UP000233060 UP000028761 UP000008225 UP000001646 UP000081671 UP000007267 UP000189706 UP000001073 UP000242457 UP000273346 UP000009130 UP000000589 UP000189704 UP000233080 UP000075880 UP000264840 UP000233180 UP000005207 UP000075883 UP000005447 UP000007646 UP000008143 UP000186698 UP000053105 UP000291020 UP000008792 UP000002494 UP000235965 UP000240080 UP000002277 UP000001070 UP000248484 UP000233120 UP000265300 UP000001519 UP000248483 UP000009136 UP000005640 UP000242638 UP000002281 UP000008227 UP000075840 UP000252040
UP000000304 UP000001292 UP000002358 UP000008711 UP000030765 UP000000803 UP000027135 UP000092553 UP000192221 UP000075900 UP000078200 UP000037069 UP000002282 UP000075886 UP000007266 UP000075884 UP000001595 UP000005203 UP000233220 UP000233200 UP000248480 UP000233020 UP000233040 UP000233100 UP000233140 UP000233060 UP000028761 UP000008225 UP000001646 UP000081671 UP000007267 UP000189706 UP000001073 UP000242457 UP000273346 UP000009130 UP000000589 UP000189704 UP000233080 UP000075880 UP000264840 UP000233180 UP000005207 UP000075883 UP000005447 UP000007646 UP000008143 UP000186698 UP000053105 UP000291020 UP000008792 UP000002494 UP000235965 UP000240080 UP000002277 UP000001070 UP000248484 UP000233120 UP000265300 UP000001519 UP000248483 UP000009136 UP000005640 UP000242638 UP000002281 UP000008227 UP000075840 UP000252040
Interpro
Gene 3D
ProteinModelPortal
H9JIJ1
A0A2H1VXD7
A0A2A4JVR6
A0A2W1B6N7
A0A212EKZ2
A0A1I8N2U0
+ More
A0A1A9WTN3 A0A1I8PTX9 B4QN98 B4HL81 K7J6J5 B3NCJ9 A0A084VCY4 Q9VT21 A0A067RL00 A0A0M3QW77 A0A1W4UMC2 A0A182RQ92 W8BMC2 D3TPY2 A0A0K8W0V4 A0A1A9UFZ0 A0A0L0C8M1 B4PEE8 A0A182Q0W1 Q641F7 A0A250XZF9 A0A034W0S0 D2A2T9 A0A182N574 H2PEM6 A0A088AV72 A0A2K6SY51 A0A0A9Z9H3 A0A2K6QXE7 A0A0K8T5L8 U5EYW6 A0A2Y9DVV3 A0A2K5CX43 A0A2K5SDA6 A0A2K5U0U8 A0A2K6A6Y4 A0A2K5MFH5 H9EVU2 A0A096N788 F7IL08 G1KSN5 A0A1S3GAL8 K7GBA2 G7NSZ6 A0A1U7QL23 G1R3M8 A0A2A3ETC7 Q3UB60 A0A3L7IHP4 G7P6H6 I7GL27 Q9CR16 A0A1U7SYN1 A0A2K5I5F7 A0A2K5UHD9 A0A182J317 A0A3Q2US37 A0A2K6KDJ4 I3IY94 A0A182MNW8 H0UVB7 G3TDA2 L5MLJ1 A0A1Z5L541 Q6P894 A0A1L8HUD3 A0A0M8ZTM2 A0A452J5W3 B4LHR5 Q6DGG0 A0A2J7PGH4 K9K4R4 S7MJ67 A0A2R9A916 H2QQC9 B4IY92 A0A2Y9F907 A0A2K6DSW2 A0A340WDL9 A0A1A7XW11 G3RAF0 A0A2Y9NSC0 A0A2K6QXE6 P26882 E5KN55 Q08752 A0A3P9NPY5 L8HPW4 F7DFM9 F1RTY6 A0A182HJD1 Q6FGM6 E5KN59 A0A341BJF3
A0A1A9WTN3 A0A1I8PTX9 B4QN98 B4HL81 K7J6J5 B3NCJ9 A0A084VCY4 Q9VT21 A0A067RL00 A0A0M3QW77 A0A1W4UMC2 A0A182RQ92 W8BMC2 D3TPY2 A0A0K8W0V4 A0A1A9UFZ0 A0A0L0C8M1 B4PEE8 A0A182Q0W1 Q641F7 A0A250XZF9 A0A034W0S0 D2A2T9 A0A182N574 H2PEM6 A0A088AV72 A0A2K6SY51 A0A0A9Z9H3 A0A2K6QXE7 A0A0K8T5L8 U5EYW6 A0A2Y9DVV3 A0A2K5CX43 A0A2K5SDA6 A0A2K5U0U8 A0A2K6A6Y4 A0A2K5MFH5 H9EVU2 A0A096N788 F7IL08 G1KSN5 A0A1S3GAL8 K7GBA2 G7NSZ6 A0A1U7QL23 G1R3M8 A0A2A3ETC7 Q3UB60 A0A3L7IHP4 G7P6H6 I7GL27 Q9CR16 A0A1U7SYN1 A0A2K5I5F7 A0A2K5UHD9 A0A182J317 A0A3Q2US37 A0A2K6KDJ4 I3IY94 A0A182MNW8 H0UVB7 G3TDA2 L5MLJ1 A0A1Z5L541 Q6P894 A0A1L8HUD3 A0A0M8ZTM2 A0A452J5W3 B4LHR5 Q6DGG0 A0A2J7PGH4 K9K4R4 S7MJ67 A0A2R9A916 H2QQC9 B4IY92 A0A2Y9F907 A0A2K6DSW2 A0A340WDL9 A0A1A7XW11 G3RAF0 A0A2Y9NSC0 A0A2K6QXE6 P26882 E5KN55 Q08752 A0A3P9NPY5 L8HPW4 F7DFM9 F1RTY6 A0A182HJD1 Q6FGM6 E5KN59 A0A341BJF3
PDB
1IIP
E-value=3.88417e-70,
Score=673
Ontologies
GO
GO:0003755
GO:0006457
GO:0051082
GO:0016018
GO:0042026
GO:0065003
GO:0043065
GO:0034389
GO:0045070
GO:0008134
GO:0000122
GO:0050714
GO:0051879
GO:0061077
GO:0005730
GO:0071492
GO:0005829
GO:0005528
GO:0005634
GO:0005654
GO:0005737
GO:0030544
GO:0005739
GO:0030331
GO:0031072
GO:0000413
GO:0015031
GO:0019899
GO:0006915
GO:0043066
GO:0006979
GO:0051881
GO:0019076
GO:0005515
GO:0019012
GO:0051603
GO:0030145
GO:0006412
GO:0016876
GO:0043039
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleolus
Nucleoplasm
Nucleus
Nucleolus
Nucleoplasm
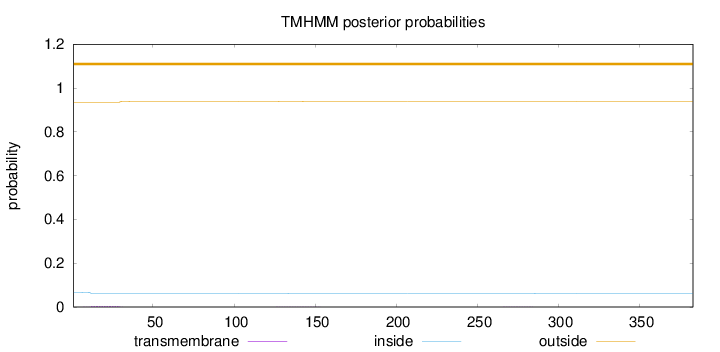
Length:
383
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.09648
Exp number, first 60 AAs:
0.0792999999999999
Total prob of N-in:
0.06601
outside
1 - 383
Population Genetic Test Statistics
Pi
234.140133
Theta
176.753619
Tajima's D
1.272581
CLR
0.182658
CSRT
0.733363331833408
Interpretation
Uncertain
