Pre Gene Modal
BGIBMGA009342
Annotation
PREDICTED:_GPI_mannosyltransferase_3_[Amyelois_transitella]
Full name
Mannosyltransferase
+ More
GPI mannosyltransferase 3
Succinate--CoA ligase [ADP/GDP-forming] subunit alpha, mitochondrial
GPI mannosyltransferase 3
Succinate--CoA ligase [ADP/GDP-forming] subunit alpha, mitochondrial
Alternative Name
GPI mannosyltransferase III
Phosphatidylinositol-glycan biosynthesis class B protein
Succinyl-CoA synthetase subunit alpha
Phosphatidylinositol-glycan biosynthesis class B protein
Succinyl-CoA synthetase subunit alpha
Location in the cell
PlasmaMembrane Reliability : 4.55
Sequence
CDS
ATGGTGTTGGCAAAGGGTTTGAGGCCCGTTCAGGTGGTCTCTTTTATATTATTCGTGAGAATACTTTCTGTATTCTTAGTGAGAACTTGGTATGTTCCTGACGAGTACTGGCAGACCTTGGAAGTGGCCCATAAACAAGTGTTCCACTATGGAGAACTCACCTGGGAATGGCAGAGGGGTATCAGGAGCTACTTATTCCCGAGCGTAGTTGCTGTCTTGTATAGTATATTGAAGGTTACTGGCTTGGATCACCCTGAAGCTGTGATTCTTCTACCTAGGATCCTACAAGCAATACTCAGTACTGCAGCAGACTACAGTTTCTATAAATGGACAGGAGGTCGGAAGTGGGCTCTGTTCCTCATTCTCACATCTTGGTTCTGGTTCTATACTTCAGGCAGAACACTGCTCCAGACTACAGAGACAGCTCTTGTGGCAATCGCCTTATCTGTTTTTCCATTCAAGAGTGGAAAACTTGGATATTATGATAAAGAGAACACATCATGGATATGGCTAGCAGTCATAGCTGTGTTTCTTCGGCCAACTTCTGCACCTCTATGGGCAGTGCTCACTATCTACAACTTACTGACAACGAACCAGTCCAAGTTGCAGTTACTGTTGAGATCATATTTACCTATTGGATTGATCGCTGGTAGTGCTTTAGTGGGTTTAGATACATACTTCTACGGAAGAGTTGTGATCACTCCATGGGAATTCTTCAAGTTTAATGTGCTTTATGATATTGCATCTTTCTATGGAAAACATCCATGGCATTGGTACTTGAGCCAGGGTCTGCCGGCCGTGCTGGGCGTCAACACAGTGCCGGTGCTGTGGGCAGCGGTCAACATAGTTCGTCGACCCCGGGAGAACAAGACGGGGGCGCTGCTGTTGTTTGCTGCAGCTCTACACATAGCACTATATAGCTTCATCTCGCACAAGGAGTTCAGATTCGTATTACCGTTACTGCCCATCCTGCTGTACCTCGCGCAGGACGCCATCGTACCGTGGAGCAGGAAAGCTAAAAGATGGCAGCTGTACGTGGTGGCGGCGTGCATGGTGGTGGGCAACGCGCTGCCGGCCGCGTACTTCGGCGCCGTGCACCAGAGGGGCGCGCTGGACGTGATGCCGCTGCTGCGGGACGCCGCCTCCACCAACCGCTCCTCCATCGCCTTCCTCATGCCCTGCCACTCGACGCCGCTCTATAGCCATATCCACAAAAACATCACAACCCGTTATCTCCACTGTGAGCCTCCGTTCAACAAGCCTGGCGAAACTTACGAATCGGAAGCCTTCTTCAACAACCCTCTCCGGTGGTGGAGGAATGAGTACTCCAGTAGACAGACACCAACTCTGGTCGTACTCTTCGATACCCTTAAAGGGAGAGTAGAAAATATTCTGAGTGGATATCGGTTGTTGCATGAGATTCCTCATACACAGTTCCCAGAAGGAGAGGTCGGTGAGAATGTCTTGGTGTATCAGAAAATAGAAAGTCAACCTAAACCGCCGGCTGACGAAGTTGTTTAA
Protein
MVLAKGLRPVQVVSFILFVRILSVFLVRTWYVPDEYWQTLEVAHKQVFHYGELTWEWQRGIRSYLFPSVVAVLYSILKVTGLDHPEAVILLPRILQAILSTAADYSFYKWTGGRKWALFLILTSWFWFYTSGRTLLQTTETALVAIALSVFPFKSGKLGYYDKENTSWIWLAVIAVFLRPTSAPLWAVLTIYNLLTTNQSKLQLLLRSYLPIGLIAGSALVGLDTYFYGRVVITPWEFFKFNVLYDIASFYGKHPWHWYLSQGLPAVLGVNTVPVLWAAVNIVRRPRENKTGALLLFAAALHIALYSFISHKEFRFVLPLLPILLYLAQDAIVPWSRKAKRWQLYVVAACMVVGNALPAAYFGAVHQRGALDVMPLLRDAASTNRSSIAFLMPCHSTPLYSHIHKNITTRYLHCEPPFNKPGETYESEAFFNNPLRWWRNEYSSRQTPTLVVLFDTLKGRVENILSGYRLLHEIPHTQFPEGEVGENVLVYQKIESQPKPPADEVV
Summary
Description
Mannosyltransferase involved in glycosylphosphatidylinositol-anchor biosynthesis. Transfers the third alpha-1,2-mannose to Man2-GlcN-acyl-PI during GPI precursor assembly (By similarity).
Succinyl-CoA synthetase functions in the citric acid cycle (TCA), coupling the hydrolysis of succinyl-CoA to the synthesis of either ATP or GTP and thus represents the only step of substrate-level phosphorylation in the TCA. The alpha subunit of the enzyme binds the substrates coenzyme A and phosphate, while succinate binding and specificity for either ATP or GTP is provided by different beta subunits.
Succinyl-CoA synthetase functions in the citric acid cycle (TCA), coupling the hydrolysis of succinyl-CoA to the synthesis of either ATP or GTP and thus represents the only step of substrate-level phosphorylation in the TCA. The alpha subunit of the enzyme binds the substrates coenzyme A and phosphate, while succinate binding and specificity for either ATP or GTP is provided by different beta subunits.
Catalytic Activity
ATP + CoA + succinate = ADP + phosphate + succinyl-CoA
CoA + GTP + succinate = GDP + phosphate + succinyl-CoA
CoA + GTP + succinate = GDP + phosphate + succinyl-CoA
Subunit
Heterodimer of an alpha and a beta subunit. Different beta subunits determine nucleotide specificity. Together with an ATP-specific beta subunit, forms an ADP-forming succinyl-CoA synthetase (A-SCS). Together with a GTP-specific beta subunit forms a GDP-forming succinyl-CoA synthetase (G-SCS).
Similarity
Belongs to the glycosyltransferase 22 family.
Belongs to the glycosyltransferase 22 family. PIGB subfamily.
Belongs to the succinate/malate CoA ligase alpha subunit family.
Belongs to the glycosyltransferase 22 family. PIGB subfamily.
Belongs to the succinate/malate CoA ligase alpha subunit family.
Keywords
Alternative splicing
Complete proteome
Endoplasmic reticulum
Glycoprotein
Glycosyltransferase
GPI-anchor biosynthesis
Membrane
Reference proteome
Transferase
Transmembrane
Transmembrane helix
Feature
chain Mannosyltransferase
splice variant In isoform B.
splice variant In isoform B.
Uniprot
H9JIJ2
A0A2A4JIR7
A0A2W1B923
A0A2H1VCT5
A0A194Q788
A0A212EKY4
+ More
A0A437B735 B4N589 A0A1W4W3Z6 B4HTU5 B3NBZ5 A0A0J9RNI0 B4PH62 B4IZW1 B3M806 B4KYA0 Q9VZM5-2 B4LDG7 A0A1J1ILD4 A0A0J9RNZ5 D6WD71 A0A0Q5UI06 A0A0R1E0S1 A0A1W4VSL4 A0A0P8XU98 A0A182GC61 A0A336MML2 A0A336MH85 Q2LZU0 A0A3B0JS25 B0X524 A0A023EUF1 A0A182M423 A0A0Q9XMV0 A0A0Q9XNI0 A0A1I8M585 A0A0K8VE28 Q9VZM5 A0A0A1X6J3 A0A0A1WQE4 W8C1D2 A0A1Q3FML2 A0A0R3P861 A0A1W4XBQ4 A0A0Q9WHY3 A0A182JVZ1 A0A182XXE7 A0A340TB88 A0A1Y9J0T6 Q7QFX4 A0A182P793 A0A1I8P309 A0A182LAQ6 A0A182SM66 A0A182I8U0 A0A1I8M584 A0A0L0CCX1 Q0C786 A0A1S4EUN4 A0A182R5W5 A0A182PZW1 A0A182N4E5 A0A182UQ54 A0A084VDC9 A0A1B0AJC0 A0A1B0BAR7 A0A1A9XJY7 A0A182FSD8 A0A1B0GNQ9 A0A1Y1NEB8 W5JB23 A0A1I8P354 A0A1B0C863 A0A1B0GF61 A0A1A9US15 B4QPY6 A0A0M4EIX9 B4H818 A0A182JMN6 A0A182UC57 A0A1Q3FLJ7 U4U3H7 A0A1A8H8T8 A0A1A8DLM9 A0A1A8JYL1 E9JD33 A0A1A8A0E2 A0A1A8PRD7 A0A1A8NJD5 A0A2A3EMT0 A0A2I4CTF0 A0A1W4XBH1 A7RSA4 Q7SXZ1 A0A0R4IFP6 W4XVD5
A0A437B735 B4N589 A0A1W4W3Z6 B4HTU5 B3NBZ5 A0A0J9RNI0 B4PH62 B4IZW1 B3M806 B4KYA0 Q9VZM5-2 B4LDG7 A0A1J1ILD4 A0A0J9RNZ5 D6WD71 A0A0Q5UI06 A0A0R1E0S1 A0A1W4VSL4 A0A0P8XU98 A0A182GC61 A0A336MML2 A0A336MH85 Q2LZU0 A0A3B0JS25 B0X524 A0A023EUF1 A0A182M423 A0A0Q9XMV0 A0A0Q9XNI0 A0A1I8M585 A0A0K8VE28 Q9VZM5 A0A0A1X6J3 A0A0A1WQE4 W8C1D2 A0A1Q3FML2 A0A0R3P861 A0A1W4XBQ4 A0A0Q9WHY3 A0A182JVZ1 A0A182XXE7 A0A340TB88 A0A1Y9J0T6 Q7QFX4 A0A182P793 A0A1I8P309 A0A182LAQ6 A0A182SM66 A0A182I8U0 A0A1I8M584 A0A0L0CCX1 Q0C786 A0A1S4EUN4 A0A182R5W5 A0A182PZW1 A0A182N4E5 A0A182UQ54 A0A084VDC9 A0A1B0AJC0 A0A1B0BAR7 A0A1A9XJY7 A0A182FSD8 A0A1B0GNQ9 A0A1Y1NEB8 W5JB23 A0A1I8P354 A0A1B0C863 A0A1B0GF61 A0A1A9US15 B4QPY6 A0A0M4EIX9 B4H818 A0A182JMN6 A0A182UC57 A0A1Q3FLJ7 U4U3H7 A0A1A8H8T8 A0A1A8DLM9 A0A1A8JYL1 E9JD33 A0A1A8A0E2 A0A1A8PRD7 A0A1A8NJD5 A0A2A3EMT0 A0A2I4CTF0 A0A1W4XBH1 A7RSA4 Q7SXZ1 A0A0R4IFP6 W4XVD5
EC Number
2.4.1.-
6.2.1.4
6.2.1.5
6.2.1.4
6.2.1.5
Pubmed
19121390
28756777
26354079
22118469
17994087
22936249
+ More
17550304 10731132 12537572 18362917 19820115 26483478 15632085 24945155 18057021 25315136 25830018 24495485 25244985 12364791 14747013 17210077 20966253 26108605 17510324 24438588 28004739 20920257 23761445 23537049 21282665 17615350 23594743
17550304 10731132 12537572 18362917 19820115 26483478 15632085 24945155 18057021 25315136 25830018 24495485 25244985 12364791 14747013 17210077 20966253 26108605 17510324 24438588 28004739 20920257 23761445 23537049 21282665 17615350 23594743
EMBL
BABH01027083
BABH01027084
NWSH01001280
PCG71861.1
KZ150210
PZC72209.1
+ More
ODYU01001865 SOQ38660.1 KQ459337 KPJ01407.1 AGBW02014173 OWR42139.1 RSAL01000133 RVE46326.1 CH964101 EDW79528.1 CH480817 EDW50366.1 CH954178 EDV50812.1 CM002912 KMY97423.1 CM000159 EDW93299.1 CH916366 EDV96733.1 CH902618 EDV39914.1 CH933809 EDW17679.1 AE014296 BT003240 CH940647 EDW68905.1 CVRI01000050 CRK99265.1 KMY97422.1 KQ971312 EEZ98306.1 KQS43526.1 KRK01092.1 KPU78291.1 JXUM01053700 JXUM01053701 JXUM01053702 JXUM01053703 KQ561787 KXJ77539.1 UFQT01001210 SSX29517.1 UFQT01001263 SSX29752.1 CH379069 EAL29416.2 OUUW01000009 SPP84917.1 DS232362 EDS40670.1 GAPW01001026 JAC12572.1 AXCM01002368 KRG05687.1 KRG05688.1 GDHF01015212 GDHF01001253 JAI37102.1 JAI51061.1 GBXI01007957 JAD06335.1 GBXI01013023 JAD01269.1 GAMC01007015 GAMC01007014 JAB99541.1 GFDL01006283 JAV28762.1 KRT07955.1 KRF84067.1 AAAB01008844 EAA06057.4 APCN01003119 JRES01000577 KNC30095.1 CH477186 EAT48940.1 AXCN02000039 ATLV01011259 KE524659 KFB35973.1 JXJN01011133 AJVK01029700 GEZM01005566 JAV96089.1 ADMH02001999 ETN60045.1 AJWK01000216 AJWK01000217 CCAG010012763 CM000363 EDX09106.1 CP012525 ALC43901.1 CH479220 EDW34812.1 GFDL01006643 JAV28402.1 KB631874 ERL86863.1 HAEC01010909 SBQ79125.1 HAEA01005958 SBQ34438.1 HAEE01005071 SBR25091.1 GL771866 EFZ09140.1 HADY01009501 HAEJ01004928 SBP47986.1 HAEH01008262 SBR84000.1 HAEG01003284 SBR68927.1 KZ288206 PBC33065.1 DS469533 EDO45709.1 BC055191 CABZ01069901 CU627990 AAGJ04037456 AAGJ04037457
ODYU01001865 SOQ38660.1 KQ459337 KPJ01407.1 AGBW02014173 OWR42139.1 RSAL01000133 RVE46326.1 CH964101 EDW79528.1 CH480817 EDW50366.1 CH954178 EDV50812.1 CM002912 KMY97423.1 CM000159 EDW93299.1 CH916366 EDV96733.1 CH902618 EDV39914.1 CH933809 EDW17679.1 AE014296 BT003240 CH940647 EDW68905.1 CVRI01000050 CRK99265.1 KMY97422.1 KQ971312 EEZ98306.1 KQS43526.1 KRK01092.1 KPU78291.1 JXUM01053700 JXUM01053701 JXUM01053702 JXUM01053703 KQ561787 KXJ77539.1 UFQT01001210 SSX29517.1 UFQT01001263 SSX29752.1 CH379069 EAL29416.2 OUUW01000009 SPP84917.1 DS232362 EDS40670.1 GAPW01001026 JAC12572.1 AXCM01002368 KRG05687.1 KRG05688.1 GDHF01015212 GDHF01001253 JAI37102.1 JAI51061.1 GBXI01007957 JAD06335.1 GBXI01013023 JAD01269.1 GAMC01007015 GAMC01007014 JAB99541.1 GFDL01006283 JAV28762.1 KRT07955.1 KRF84067.1 AAAB01008844 EAA06057.4 APCN01003119 JRES01000577 KNC30095.1 CH477186 EAT48940.1 AXCN02000039 ATLV01011259 KE524659 KFB35973.1 JXJN01011133 AJVK01029700 GEZM01005566 JAV96089.1 ADMH02001999 ETN60045.1 AJWK01000216 AJWK01000217 CCAG010012763 CM000363 EDX09106.1 CP012525 ALC43901.1 CH479220 EDW34812.1 GFDL01006643 JAV28402.1 KB631874 ERL86863.1 HAEC01010909 SBQ79125.1 HAEA01005958 SBQ34438.1 HAEE01005071 SBR25091.1 GL771866 EFZ09140.1 HADY01009501 HAEJ01004928 SBP47986.1 HAEH01008262 SBR84000.1 HAEG01003284 SBR68927.1 KZ288206 PBC33065.1 DS469533 EDO45709.1 BC055191 CABZ01069901 CU627990 AAGJ04037456 AAGJ04037457
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000283053
UP000007798
+ More
UP000192221 UP000001292 UP000008711 UP000002282 UP000001070 UP000007801 UP000009192 UP000000803 UP000008792 UP000183832 UP000007266 UP000069940 UP000249989 UP000001819 UP000268350 UP000002320 UP000075883 UP000095301 UP000192223 UP000075881 UP000076408 UP000075920 UP000076407 UP000007062 UP000075885 UP000095300 UP000075882 UP000075901 UP000075840 UP000037069 UP000008820 UP000075900 UP000075886 UP000075884 UP000075903 UP000030765 UP000092445 UP000092460 UP000092443 UP000069272 UP000092462 UP000000673 UP000092461 UP000092444 UP000078200 UP000000304 UP000092553 UP000008744 UP000075880 UP000075902 UP000030742 UP000242457 UP000192220 UP000001593 UP000000437 UP000007110
UP000192221 UP000001292 UP000008711 UP000002282 UP000001070 UP000007801 UP000009192 UP000000803 UP000008792 UP000183832 UP000007266 UP000069940 UP000249989 UP000001819 UP000268350 UP000002320 UP000075883 UP000095301 UP000192223 UP000075881 UP000076408 UP000075920 UP000076407 UP000007062 UP000075885 UP000095300 UP000075882 UP000075901 UP000075840 UP000037069 UP000008820 UP000075900 UP000075886 UP000075884 UP000075903 UP000030765 UP000092445 UP000092460 UP000092443 UP000069272 UP000092462 UP000000673 UP000092461 UP000092444 UP000078200 UP000000304 UP000092553 UP000008744 UP000075880 UP000075902 UP000030742 UP000242457 UP000192220 UP000001593 UP000000437 UP000007110
Pfam
Interpro
IPR039521
PIG-B/GPI10
+ More
IPR005599 GPI_mannosylTrfase
IPR027417 P-loop_NTPase
IPR013632 DNA_recomb/repair_Rad51_C
IPR020588 RecA_ATP-bd
IPR003593 AAA+_ATPase
IPR036291 NAD(P)-bd_dom_sf
IPR005811 CoA_ligase
IPR003781 CoA-bd
IPR005810 CoA_lig_alpha
IPR033847 Citrt_syn/SCS-alpha_CS
IPR017440 Cit_synth/succinyl-CoA_lig_AS
IPR016102 Succinyl-CoA_synth-like
IPR019018 Rab-bd_FIP-RBD
IPR037245 FIP-RBD_C_sf
IPR005599 GPI_mannosylTrfase
IPR027417 P-loop_NTPase
IPR013632 DNA_recomb/repair_Rad51_C
IPR020588 RecA_ATP-bd
IPR003593 AAA+_ATPase
IPR036291 NAD(P)-bd_dom_sf
IPR005811 CoA_ligase
IPR003781 CoA-bd
IPR005810 CoA_lig_alpha
IPR033847 Citrt_syn/SCS-alpha_CS
IPR017440 Cit_synth/succinyl-CoA_lig_AS
IPR016102 Succinyl-CoA_synth-like
IPR019018 Rab-bd_FIP-RBD
IPR037245 FIP-RBD_C_sf
Gene 3D
ProteinModelPortal
H9JIJ2
A0A2A4JIR7
A0A2W1B923
A0A2H1VCT5
A0A194Q788
A0A212EKY4
+ More
A0A437B735 B4N589 A0A1W4W3Z6 B4HTU5 B3NBZ5 A0A0J9RNI0 B4PH62 B4IZW1 B3M806 B4KYA0 Q9VZM5-2 B4LDG7 A0A1J1ILD4 A0A0J9RNZ5 D6WD71 A0A0Q5UI06 A0A0R1E0S1 A0A1W4VSL4 A0A0P8XU98 A0A182GC61 A0A336MML2 A0A336MH85 Q2LZU0 A0A3B0JS25 B0X524 A0A023EUF1 A0A182M423 A0A0Q9XMV0 A0A0Q9XNI0 A0A1I8M585 A0A0K8VE28 Q9VZM5 A0A0A1X6J3 A0A0A1WQE4 W8C1D2 A0A1Q3FML2 A0A0R3P861 A0A1W4XBQ4 A0A0Q9WHY3 A0A182JVZ1 A0A182XXE7 A0A340TB88 A0A1Y9J0T6 Q7QFX4 A0A182P793 A0A1I8P309 A0A182LAQ6 A0A182SM66 A0A182I8U0 A0A1I8M584 A0A0L0CCX1 Q0C786 A0A1S4EUN4 A0A182R5W5 A0A182PZW1 A0A182N4E5 A0A182UQ54 A0A084VDC9 A0A1B0AJC0 A0A1B0BAR7 A0A1A9XJY7 A0A182FSD8 A0A1B0GNQ9 A0A1Y1NEB8 W5JB23 A0A1I8P354 A0A1B0C863 A0A1B0GF61 A0A1A9US15 B4QPY6 A0A0M4EIX9 B4H818 A0A182JMN6 A0A182UC57 A0A1Q3FLJ7 U4U3H7 A0A1A8H8T8 A0A1A8DLM9 A0A1A8JYL1 E9JD33 A0A1A8A0E2 A0A1A8PRD7 A0A1A8NJD5 A0A2A3EMT0 A0A2I4CTF0 A0A1W4XBH1 A7RSA4 Q7SXZ1 A0A0R4IFP6 W4XVD5
A0A437B735 B4N589 A0A1W4W3Z6 B4HTU5 B3NBZ5 A0A0J9RNI0 B4PH62 B4IZW1 B3M806 B4KYA0 Q9VZM5-2 B4LDG7 A0A1J1ILD4 A0A0J9RNZ5 D6WD71 A0A0Q5UI06 A0A0R1E0S1 A0A1W4VSL4 A0A0P8XU98 A0A182GC61 A0A336MML2 A0A336MH85 Q2LZU0 A0A3B0JS25 B0X524 A0A023EUF1 A0A182M423 A0A0Q9XMV0 A0A0Q9XNI0 A0A1I8M585 A0A0K8VE28 Q9VZM5 A0A0A1X6J3 A0A0A1WQE4 W8C1D2 A0A1Q3FML2 A0A0R3P861 A0A1W4XBQ4 A0A0Q9WHY3 A0A182JVZ1 A0A182XXE7 A0A340TB88 A0A1Y9J0T6 Q7QFX4 A0A182P793 A0A1I8P309 A0A182LAQ6 A0A182SM66 A0A182I8U0 A0A1I8M584 A0A0L0CCX1 Q0C786 A0A1S4EUN4 A0A182R5W5 A0A182PZW1 A0A182N4E5 A0A182UQ54 A0A084VDC9 A0A1B0AJC0 A0A1B0BAR7 A0A1A9XJY7 A0A182FSD8 A0A1B0GNQ9 A0A1Y1NEB8 W5JB23 A0A1I8P354 A0A1B0C863 A0A1B0GF61 A0A1A9US15 B4QPY6 A0A0M4EIX9 B4H818 A0A182JMN6 A0A182UC57 A0A1Q3FLJ7 U4U3H7 A0A1A8H8T8 A0A1A8DLM9 A0A1A8JYL1 E9JD33 A0A1A8A0E2 A0A1A8PRD7 A0A1A8NJD5 A0A2A3EMT0 A0A2I4CTF0 A0A1W4XBH1 A7RSA4 Q7SXZ1 A0A0R4IFP6 W4XVD5
Ontologies
PATHWAY
GO
PANTHER
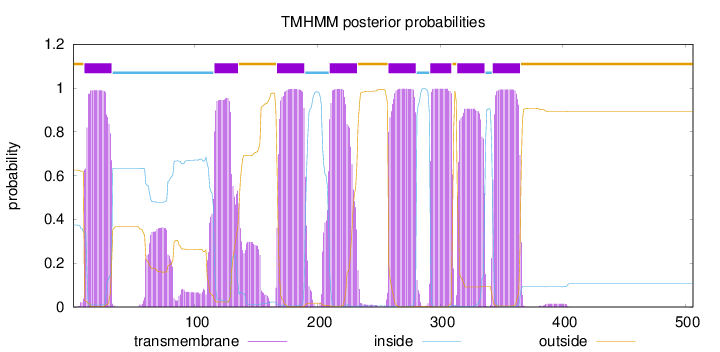
Topology
Subcellular location
Endoplasmic reticulum membrane
Mitochondrion
Mitochondrion
Length:
506
Number of predicted TMHs:
8
Exp number of AAs in TMHs:
182.4978
Exp number, first 60 AAs:
21.35114
Total prob of N-in:
0.37411
POSSIBLE N-term signal
sequence
outside
1 - 9
TMhelix
10 - 32
inside
33 - 115
TMhelix
116 - 135
outside
136 - 166
TMhelix
167 - 189
inside
190 - 209
TMhelix
210 - 232
outside
233 - 257
TMhelix
258 - 280
inside
281 - 291
TMhelix
292 - 309
outside
310 - 313
TMhelix
314 - 336
inside
337 - 342
TMhelix
343 - 365
outside
366 - 506
Population Genetic Test Statistics
Pi
200.827968
Theta
201.268967
Tajima's D
0.475086
CLR
0.20644
CSRT
0.510724463776811
Interpretation
Uncertain
