Gene
KWMTBOMO08497
Pre Gene Modal
BGIBMGA009343
Annotation
PREDICTED:_hemicentin-1-like_isoform_X1_[Bombyx_mori]
Full name
Hemicentin-1
Alternative Name
Fibulin-6
Location in the cell
Extracellular Reliability : 2.588
Sequence
CDS
ATGTTTCATTCCAGTTTCAATGACTACGGTAATCTGTACATAGTGAACGCTAGTGTCCAAACAGAAGGAGTTTACAGTTGCATCGCTGGTAACATCGCCGGTGTTGCCGTGAAGACGATTTTCCTTAGCGTTAATGCACCACCGAAAATTCTAGAAGATAATTATACCGGCTCGTATGTGGCCACCGATATGGATCATTCATTGACAATATATTGTTCCGCAACCGGCAAACCTAAGCCGTATGTCCTTTGGTCTAAGGACGAGTTTTATTTGAATCACGATCCTCGTTACGACGTGGACTTCAATGGTAGGCTAACGATACAGTCGCCCGCCGAGGAGCTAAGCGGAAACTACACTTGCGAAGCCAGAAACAAGTTCGGGTATACGAACAAGACAGTCGAAGTAAACATTTATTCCGTGCCGAAACAAGTACAATCAGCCGAAAGCTATGCGAAGGTGACAGCACTCGAAGGAACCGACATTGTCATCGCGTGTCCCATCCGAGCTACAAACGTGCGATCAATATCATGGTACAAGGAAGCAAAATTAATATCAAAAGGTGATCTTCAGCTAAAAAACGTTTCGCGAAATGACACGTCCGTGTACGCGTGCGTCGCGACGAACACTGCGGGCAGCGCGCACGCGTCCGTAGCGCTCGACGTGCAATGGCCGCCCGCGTTCGTGACGAGGGTAGACGGAAATATTGAGGTCATCAAAGGAGAGGACGCGTGGTTCAGCTGTGCAGTCGACGCTAAACCTTTGGCTAAGATAAAATGGTTATTCAATTCTAAGCCATTGATCTTCGAGGATAAGGATGTTTTTAAAGTATTAAACGCACAACAAAGAAACGCGGGCGTTTATAAATGTGTTGTTAGCAACATTCATGGAACTATAATCAAGGCGTTCAAATTGTCCGTCCTGGAGCCTCCATACCTAATGGAGTTTGACTTTTTGGATGTATTGCTGAAGGAAGGGAGTAATGCTACACTTGAGTGTGTGGCTAAAGGAGTTCCCGCACCGACTGTCCAATGGATCTACAACAACACTCAGTGGGTGGCAGACGGTCCAGCCATAACCAGCACGAATACAACAACGGAAGCGGAAGGCCTGTATCGGTGCGAGGCTGCGAACAAAGCCGGAAGCGCGCACGTGGTCTACAGGGTGGTGGTGCTCGCGGCGCCGTCTGTGCTCCACGTCACGTCCTACAGCGACGGACGAGGGAGCACCGTCGACGAAGCCGTCGAAGTTATTCTCAACAAACGTGCCAGAATATCGTGTAAAGCAAAAGGGAGACCGGAACCCATCATACAGTGGTTCAAGAACGGTCGCGTCGTTGGTCAAAATCTACCTGGAATTAGCAGCGCCGATGTGGTACTGGATAGCGTGCAGGCCTCACATGCCGGGGCCTACACGTGTGTCGCCACTAATGAAGGCGGTAGTGATCAGAAGCGAATCAGACTTGAGGTCTTAGAAAAACCAAACATATTCCGCACTATTTTCCAAAGCGAAAACATAACGAGTAACACGATTAAATTAGACGTGTTAGTGGGGCAAGCGTTTTATATCCACTGCCACCCGACGGGCAGCCCGCCGCCGGACATCTACTGGTTCAAAGACGGGCTCCCGCTGATGCTGTACGATGACACTATGGTCTCGACTGAATTCAACGAGGTCTTAGTCTCGAAGGCCGCTCTGGAGGACCAGGCCGGGAACTATACTTGCATCGTGAGGAATAAGGTTGGCAATAACAGCGTCAATTATCTAGTCGACGTCTTAGTGCCTCCGCCAATACAGAAAGAGAAAACGAAACGGGTGTCAGCTCGACTGGGAATGTCTGTGGTATTGACATGTCCGGTGGAGGGCAGTCCCCTACCAGACGTGATGTGGATTAAACACCCTTACACCGAAATAGCCGAGGGCTCTGATAAAATGTCGCTCTCTGATGACAATGTTACTCTGATAATAAACAACACTGACGTATCGGATACCGGGAAATACTCGTGCGTTATGACAAACAAGGTTGGCACCACTGAAGTTGTGTTCGACGTGATCATATTGAAGGCGCCGTCCATTGCCGCTAATGTAGGAGGTAGCGCGATAGAAAGACACGTTGTACCATTGAAAAGGAGCGTTGTACTCAAATGCGAAGCGGACGGTCACCCGACACCGAAAATTACCTGGTTAAAGGACACGCAGCGTCTGAGCGAGAGTGCGTCGAACGTCGTGCGCGTCCTCGGCAACAGCTTGCTCTACGTCCGGGGCGGGTCCTCCCGCGACGCCGGCCACTACATCTGCGTGGCGGAGAACGAGGCCGGCACCGCGCACCGGAGGTACAACCTCGCCGTGCAGGTTCCGGGCAAATGGAGCGATTGGTCTAAATGGTCTTACTGCAACGTCACGTGCGGCCCTGGATACCAGTTCCGGTCCAGATCGTGTCACTACATCGACGAATACAACAACACCATAGATAATACAACTCAGTCGGACACGGTCATAGTTGACGAGAGCGCGTGCAAGGGAGAGAGTGGGGACAAACGAAAATGCAGGATGCCACAATGTGAGGATGAAGAGAGCCATCCGAAGTGGTCAGTGTGGTCGCGCTGGTCGCAGTGCAGCTCGAGCTGCGGCTCCGGGACGCAGACCAGGACAAGACGGTGCCGGACCAAGGTCAAATGTACCGGGGACAATGTTCAAGTTAGGAAATGTCCCGATCTACCGAAATGCGATAATACGGCGGATACGACGAGTACTTCTGACAATGACGTTTTGGACAGACATTCACACCACAACGACGATGAAGTGTATACGCCGGAGGCAGTCTACGAAATGGAACCAGAGATAATAAAATCTAAATATTCGCCTAACGTTGAACAGTTCTACGTGGCGGCGGGAACACGAGCGACGACAGCCTACTATGACGTCAACGTCACCAGCAACCTCGACCGAAGCGAGAGAGGCCGCTGCAACCCAGGGTACACGTACGAGCGGACCACGGACACGTGCCACGGTAATCTTATCTATCTCTTCGATTTGCAAAAATCGGTGAAAATTGTTGAAAAATATACGACTATCACCCCTCTAATTTTGATGGCAGAAACATTCCCTGGAGTGCCAGGCAGAGCCGGTGCTGAAGATTCATTGTAA
Protein
MFHSSFNDYGNLYIVNASVQTEGVYSCIAGNIAGVAVKTIFLSVNAPPKILEDNYTGSYVATDMDHSLTIYCSATGKPKPYVLWSKDEFYLNHDPRYDVDFNGRLTIQSPAEELSGNYTCEARNKFGYTNKTVEVNIYSVPKQVQSAESYAKVTALEGTDIVIACPIRATNVRSISWYKEAKLISKGDLQLKNVSRNDTSVYACVATNTAGSAHASVALDVQWPPAFVTRVDGNIEVIKGEDAWFSCAVDAKPLAKIKWLFNSKPLIFEDKDVFKVLNAQQRNAGVYKCVVSNIHGTIIKAFKLSVLEPPYLMEFDFLDVLLKEGSNATLECVAKGVPAPTVQWIYNNTQWVADGPAITSTNTTTEAEGLYRCEAANKAGSAHVVYRVVVLAAPSVLHVTSYSDGRGSTVDEAVEVILNKRARISCKAKGRPEPIIQWFKNGRVVGQNLPGISSADVVLDSVQASHAGAYTCVATNEGGSDQKRIRLEVLEKPNIFRTIFQSENITSNTIKLDVLVGQAFYIHCHPTGSPPPDIYWFKDGLPLMLYDDTMVSTEFNEVLVSKAALEDQAGNYTCIVRNKVGNNSVNYLVDVLVPPPIQKEKTKRVSARLGMSVVLTCPVEGSPLPDVMWIKHPYTEIAEGSDKMSLSDDNVTLIINNTDVSDTGKYSCVMTNKVGTTEVVFDVIILKAPSIAANVGGSAIERHVVPLKRSVVLKCEADGHPTPKITWLKDTQRLSESASNVVRVLGNSLLYVRGGSSRDAGHYICVAENEAGTAHRRYNLAVQVPGKWSDWSKWSYCNVTCGPGYQFRSRSCHYIDEYNNTIDNTTQSDTVIVDESACKGESGDKRKCRMPQCEDEESHPKWSVWSRWSQCSSSCGSGTQTRTRRCRTKVKCTGDNVQVRKCPDLPKCDNTADTTSTSDNDVLDRHSHHNDDEVYTPEAVYEMEPEIIKSKYSPNVEQFYVAAGTRATTAYYDVNVTSNLDRSERGRCNPGYTYERTTDTCHGNLIYLFDLQKSVKIVEKYTTITPLILMAETFPGVPGRAGAEDSL
Summary
Description
Promotes cleavage furrow maturation during cytokinesis in preimplantation embryos. May play a role in the architecture of adhesive and flexible epithelial cell junctions. May play a role during myocardial remodeling by imparting an effect on cardiac fibroblast migration.
Promotes cleavage furrow maturation during cytokinesis in preimplantation embryos (PubMed:21215633). May play a role in the architecture of adhesive and flexible epithelial cell junctions (PubMed:17015624). May play a role during myocardial remodeling by imparting an effect on cardiac fibroblast migration (PubMed:24951538).
Promotes cleavage furrow maturation during cytokinesis in preimplantation embryos (PubMed:21215633). May play a role in the architecture of adhesive and flexible epithelial cell junctions (PubMed:17015624). May play a role during myocardial remodeling by imparting an effect on cardiac fibroblast migration (PubMed:24951538).
Keywords
Age-related macular degeneration
Alternative splicing
Basement membrane
Calcium
Cell cycle
Cell division
Cell junction
Complete proteome
Cytoplasm
Disease mutation
Disulfide bond
EGF-like domain
Extracellular matrix
Glycoprotein
Immunoglobulin domain
Polymorphism
Reference proteome
Repeat
Secreted
Sensory transduction
Signal
Vision
Feature
chain Hemicentin-1
splice variant In isoform 3.
sequence variant In dbSNP:rs7539719.
splice variant In isoform 3.
sequence variant In dbSNP:rs7539719.
Uniprot
H9JIJ3
A0A2W1BAW8
A0A2A4JKA5
A0A194Q7A1
A0A3S2L5P2
A0A2H1VVZ8
+ More
A0A2H1VB14 A0A2Z5U737 A0A0N1PJS4 A0A2J7R082 A0A2J7R083 A0A1B6L2U6 A0A2Y9PQD3 A0A341BXZ8 A0A2U4CQ22 A0A2Y9TH90 A0A340WQW6 G1T5K0 Q96RW7 Q96RW7-2 A0A2J8V894 G3QH37 H2Q0S6 K7CJZ0 A0A2J8N6C6 H2N4D8 A0A1B6G7Y9 A0A287DE85 I3MFI4 A0A383YVW7 M3X1M2 F6QWR8 F1PXF9 G1MRA1 A0A2K5NVX0 A0A444UCA5 A0A3Q0CW37 A0A091V0J0 A0A2K5NWC4 A0A2K6RDI0 F7HTR8 A0A099ZVY6 A0A0D9RKF6 A0A2K6JQ42 A0A2K5ZYP4 A0A2K5K644 A0A3B3RV35 A0A2K5K656 A0A2K5TLR4 G7MF24 A0A3Q7XQ40 M3YKJ2 A0A384CL93 A0A2K5TLS7 A0A2Y9GGI7 A0A2K6BJH9 F7BH35 G3RN13 A0A2K6BJK2 A0A096NP62 F7BH08 A0A3B4GRR4 A0A1V4JZI6 A0A1V4JZF2 F6TQY1 A0A2I3MCW5 A0A3Q3GU11 A0A3P8PUT5 A0A2K6S2B6 A0A2R8ZPW9 A0A2K6S293 A0A3P9BQE6 W5M0S1 F6UCP4 V8P2G0 A0A452FS37 A0A2I3SUI4 A0A1I7U001 A0A1U7UYH7 A0A3P9NWC9 D3YXG0 F6R9N9 D3YXG0-2 A0A2U3WBH9 A0A1I7U000 A0A2I4B518 S9XZ12 A0A2K5SD05 A0A2Y9JST8 A0A3Q7T881 A0A2K5EMD4 A0A401PF95 L5KZE1 K7G868 W5P565 A0A3B4E5X1 A0A146YJD2 M7BBY2 A0A146YJH4 H0XAK9
A0A2H1VB14 A0A2Z5U737 A0A0N1PJS4 A0A2J7R082 A0A2J7R083 A0A1B6L2U6 A0A2Y9PQD3 A0A341BXZ8 A0A2U4CQ22 A0A2Y9TH90 A0A340WQW6 G1T5K0 Q96RW7 Q96RW7-2 A0A2J8V894 G3QH37 H2Q0S6 K7CJZ0 A0A2J8N6C6 H2N4D8 A0A1B6G7Y9 A0A287DE85 I3MFI4 A0A383YVW7 M3X1M2 F6QWR8 F1PXF9 G1MRA1 A0A2K5NVX0 A0A444UCA5 A0A3Q0CW37 A0A091V0J0 A0A2K5NWC4 A0A2K6RDI0 F7HTR8 A0A099ZVY6 A0A0D9RKF6 A0A2K6JQ42 A0A2K5ZYP4 A0A2K5K644 A0A3B3RV35 A0A2K5K656 A0A2K5TLR4 G7MF24 A0A3Q7XQ40 M3YKJ2 A0A384CL93 A0A2K5TLS7 A0A2Y9GGI7 A0A2K6BJH9 F7BH35 G3RN13 A0A2K6BJK2 A0A096NP62 F7BH08 A0A3B4GRR4 A0A1V4JZI6 A0A1V4JZF2 F6TQY1 A0A2I3MCW5 A0A3Q3GU11 A0A3P8PUT5 A0A2K6S2B6 A0A2R8ZPW9 A0A2K6S293 A0A3P9BQE6 W5M0S1 F6UCP4 V8P2G0 A0A452FS37 A0A2I3SUI4 A0A1I7U001 A0A1U7UYH7 A0A3P9NWC9 D3YXG0 F6R9N9 D3YXG0-2 A0A2U3WBH9 A0A1I7U000 A0A2I4B518 S9XZ12 A0A2K5SD05 A0A2Y9JST8 A0A3Q7T881 A0A2K5EMD4 A0A401PF95 L5KZE1 K7G868 W5P565 A0A3B4E5X1 A0A146YJD2 M7BBY2 A0A146YJH4 H0XAK9
Pubmed
19121390
28756777
26354079
21993624
14702039
16710414
+ More
14570714 22398555 16136131 17975172 17495919 16341006 20838655 26319212 25362486 29240929 22002653 17431167 19892987 22722832 25186727 24297900 19468303 17015624 21183079 21215633 24951538 18464734 23149746 30297745 23258410 17381049 20809919 23624526
14570714 22398555 16136131 17975172 17495919 16341006 20838655 26319212 25362486 29240929 22002653 17431167 19892987 22722832 25186727 24297900 19468303 17015624 21183079 21215633 24951538 18464734 23149746 30297745 23258410 17381049 20809919 23624526
EMBL
BABH01027071
BABH01027072
BABH01027073
BABH01027074
BABH01027075
BABH01027076
+ More
BABH01027077 BABH01027078 BABH01027079 BABH01027080 KZ150210 PZC72208.1 NWSH01001280 PCG71863.1 KQ459337 KPJ01408.1 RSAL01000133 RVE46324.1 ODYU01004773 SOQ45009.1 ODYU01001595 SOQ38038.1 AP017486 BBB06744.1 KQ459928 KPJ19296.1 NEVH01008283 PNF34248.1 PNF34243.1 GEBQ01022123 JAT17854.1 AAGW02054358 AAGW02054359 AAGW02054360 AAGW02054361 AAGW02054362 AAGW02054363 AF156100 AK056336 AK056557 AL118512 AL121996 AL133515 AL133553 AL135796 AL135797 AL391824 BX928748 AJ306906 NDHI03003429 PNJ53742.1 CABD030007797 CABD030007798 CABD030007799 CABD030007800 CABD030007801 CABD030007802 CABD030007803 CABD030007804 CABD030007805 CABD030007806 AACZ04056152 AACZ04056153 AACZ04056154 GABE01001833 NBAG03000235 JAA42906.1 PNI67324.1 PNI67325.1 ABGA01049740 ABGA01049741 ABGA01049742 ABGA01049743 ABGA01049744 ABGA01049745 ABGA01049746 ABGA01049747 ABGA01049748 ABGA01049749 GECZ01011376 JAS58393.1 AGTP01014814 AGTP01014815 AANG04001043 AAEX03005163 AAEX03005164 SCEB01214843 RXM32791.1 KL410290 KFQ95785.1 KL869911 KGL86449.1 AQIB01112956 AQIB01112957 AQIB01112958 AQIB01112959 AQIB01112960 AQIB01112961 AQIA01001031 AQIA01001032 AQIA01001033 AQIA01001034 AQIA01001035 AQIA01001036 CM001253 EHH15721.1 AEYP01068968 AEYP01068969 AEYP01068970 AEYP01068971 AEYP01068972 AEYP01068973 AEYP01068974 AEYP01068975 AEYP01068976 AEYP01068977 JSUE03003236 JSUE03003237 JSUE03003238 JSUE03003239 JSUE03003240 AHZZ02017566 AHZZ02017567 AHZZ02017568 AHZZ02017569 LSYS01005418 OPJ77523.1 OPJ77524.1 AJFE02052069 AJFE02052070 AJFE02052071 AJFE02052072 AJFE02052073 AJFE02052074 AJFE02052075 AHAT01034683 AHAT01034684 AHAT01034685 AHAT01034686 AZIM01000955 ETE68709.1 LWLT01000016 AC111145 AC113326 AC115051 AC124120 KB017072 EPY80556.1 BFAA01000395 GCB71820.1 KB030474 ELK16173.1 AGCU01004760 AGCU01004761 AGCU01004762 AGCU01004763 AGCU01004764 AGCU01004765 AGCU01004766 AGCU01004767 AGCU01004768 AGCU01004769 AMGL01022558 AMGL01022559 AMGL01022560 AMGL01022561 AMGL01022562 AMGL01022563 AMGL01022564 AMGL01022565 AMGL01022566 AMGL01022567 GCES01032043 JAR54280.1 KB532021 EMP34679.1 GCES01032044 JAR54279.1 AAQR03037752 AAQR03037753 AAQR03037754 AAQR03037755 AAQR03037756 AAQR03037757 AAQR03037758 AAQR03037759 AAQR03037760 AAQR03037761
BABH01027077 BABH01027078 BABH01027079 BABH01027080 KZ150210 PZC72208.1 NWSH01001280 PCG71863.1 KQ459337 KPJ01408.1 RSAL01000133 RVE46324.1 ODYU01004773 SOQ45009.1 ODYU01001595 SOQ38038.1 AP017486 BBB06744.1 KQ459928 KPJ19296.1 NEVH01008283 PNF34248.1 PNF34243.1 GEBQ01022123 JAT17854.1 AAGW02054358 AAGW02054359 AAGW02054360 AAGW02054361 AAGW02054362 AAGW02054363 AF156100 AK056336 AK056557 AL118512 AL121996 AL133515 AL133553 AL135796 AL135797 AL391824 BX928748 AJ306906 NDHI03003429 PNJ53742.1 CABD030007797 CABD030007798 CABD030007799 CABD030007800 CABD030007801 CABD030007802 CABD030007803 CABD030007804 CABD030007805 CABD030007806 AACZ04056152 AACZ04056153 AACZ04056154 GABE01001833 NBAG03000235 JAA42906.1 PNI67324.1 PNI67325.1 ABGA01049740 ABGA01049741 ABGA01049742 ABGA01049743 ABGA01049744 ABGA01049745 ABGA01049746 ABGA01049747 ABGA01049748 ABGA01049749 GECZ01011376 JAS58393.1 AGTP01014814 AGTP01014815 AANG04001043 AAEX03005163 AAEX03005164 SCEB01214843 RXM32791.1 KL410290 KFQ95785.1 KL869911 KGL86449.1 AQIB01112956 AQIB01112957 AQIB01112958 AQIB01112959 AQIB01112960 AQIB01112961 AQIA01001031 AQIA01001032 AQIA01001033 AQIA01001034 AQIA01001035 AQIA01001036 CM001253 EHH15721.1 AEYP01068968 AEYP01068969 AEYP01068970 AEYP01068971 AEYP01068972 AEYP01068973 AEYP01068974 AEYP01068975 AEYP01068976 AEYP01068977 JSUE03003236 JSUE03003237 JSUE03003238 JSUE03003239 JSUE03003240 AHZZ02017566 AHZZ02017567 AHZZ02017568 AHZZ02017569 LSYS01005418 OPJ77523.1 OPJ77524.1 AJFE02052069 AJFE02052070 AJFE02052071 AJFE02052072 AJFE02052073 AJFE02052074 AJFE02052075 AHAT01034683 AHAT01034684 AHAT01034685 AHAT01034686 AZIM01000955 ETE68709.1 LWLT01000016 AC111145 AC113326 AC115051 AC124120 KB017072 EPY80556.1 BFAA01000395 GCB71820.1 KB030474 ELK16173.1 AGCU01004760 AGCU01004761 AGCU01004762 AGCU01004763 AGCU01004764 AGCU01004765 AGCU01004766 AGCU01004767 AGCU01004768 AGCU01004769 AMGL01022558 AMGL01022559 AMGL01022560 AMGL01022561 AMGL01022562 AMGL01022563 AMGL01022564 AMGL01022565 AMGL01022566 AMGL01022567 GCES01032043 JAR54280.1 KB532021 EMP34679.1 GCES01032044 JAR54279.1 AAQR03037752 AAQR03037753 AAQR03037754 AAQR03037755 AAQR03037756 AAQR03037757 AAQR03037758 AAQR03037759 AAQR03037760 AAQR03037761
Proteomes
UP000005204
UP000218220
UP000053268
UP000283053
UP000053240
UP000235965
+ More
UP000248483 UP000252040 UP000245320 UP000248484 UP000265300 UP000001811 UP000005640 UP000001519 UP000002277 UP000001595 UP000005215 UP000261681 UP000011712 UP000002280 UP000002254 UP000001645 UP000233060 UP000189706 UP000053283 UP000233200 UP000008225 UP000053858 UP000029965 UP000233180 UP000233140 UP000233080 UP000261540 UP000233100 UP000286642 UP000000715 UP000261680 UP000248481 UP000233120 UP000006718 UP000028761 UP000261460 UP000190648 UP000002281 UP000261660 UP000265100 UP000233220 UP000240080 UP000265160 UP000018468 UP000291000 UP000095282 UP000189704 UP000242638 UP000000589 UP000002279 UP000245340 UP000192220 UP000233040 UP000248482 UP000286640 UP000233020 UP000288216 UP000010552 UP000007267 UP000002356 UP000261440 UP000031443 UP000005225
UP000248483 UP000252040 UP000245320 UP000248484 UP000265300 UP000001811 UP000005640 UP000001519 UP000002277 UP000001595 UP000005215 UP000261681 UP000011712 UP000002280 UP000002254 UP000001645 UP000233060 UP000189706 UP000053283 UP000233200 UP000008225 UP000053858 UP000029965 UP000233180 UP000233140 UP000233080 UP000261540 UP000233100 UP000286642 UP000000715 UP000261680 UP000248481 UP000233120 UP000006718 UP000028761 UP000261460 UP000190648 UP000002281 UP000261660 UP000265100 UP000233220 UP000240080 UP000265160 UP000018468 UP000291000 UP000095282 UP000189704 UP000242638 UP000000589 UP000002279 UP000245340 UP000192220 UP000233040 UP000248482 UP000286640 UP000233020 UP000288216 UP000010552 UP000007267 UP000002356 UP000261440 UP000031443 UP000005225
Pfam
Interpro
IPR013783
Ig-like_fold
+ More
IPR013151 Immunoglobulin
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR013098 Ig_I-set
IPR001881 EGF-like_Ca-bd_dom
IPR036383 TSP1_rpt_sf
IPR000884 TSP1_rpt
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR036179 Ig-like_dom_sf
IPR000742 EGF-like_dom
IPR003598 Ig_sub2
IPR013032 EGF-like_CS
IPR018097 EGF_Ca-bd_CS
IPR036465 vWFA_dom_sf
IPR009030 Growth_fac_rcpt_cys_sf
IPR009017 GFP
IPR006605 G2_nidogen/fibulin_G2F
IPR013106 Ig_V-set
IPR009138 Neural_cell_adh
IPR026823 cEGF
IPR032984 Hemicentin-2
IPR016130 Tyr_Pase_AS
IPR013151 Immunoglobulin
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR013098 Ig_I-set
IPR001881 EGF-like_Ca-bd_dom
IPR036383 TSP1_rpt_sf
IPR000884 TSP1_rpt
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR036179 Ig-like_dom_sf
IPR000742 EGF-like_dom
IPR003598 Ig_sub2
IPR013032 EGF-like_CS
IPR018097 EGF_Ca-bd_CS
IPR036465 vWFA_dom_sf
IPR009030 Growth_fac_rcpt_cys_sf
IPR009017 GFP
IPR006605 G2_nidogen/fibulin_G2F
IPR013106 Ig_V-set
IPR009138 Neural_cell_adh
IPR026823 cEGF
IPR032984 Hemicentin-2
IPR016130 Tyr_Pase_AS
SUPFAM
Gene 3D
CDD
ProteinModelPortal
H9JIJ3
A0A2W1BAW8
A0A2A4JKA5
A0A194Q7A1
A0A3S2L5P2
A0A2H1VVZ8
+ More
A0A2H1VB14 A0A2Z5U737 A0A0N1PJS4 A0A2J7R082 A0A2J7R083 A0A1B6L2U6 A0A2Y9PQD3 A0A341BXZ8 A0A2U4CQ22 A0A2Y9TH90 A0A340WQW6 G1T5K0 Q96RW7 Q96RW7-2 A0A2J8V894 G3QH37 H2Q0S6 K7CJZ0 A0A2J8N6C6 H2N4D8 A0A1B6G7Y9 A0A287DE85 I3MFI4 A0A383YVW7 M3X1M2 F6QWR8 F1PXF9 G1MRA1 A0A2K5NVX0 A0A444UCA5 A0A3Q0CW37 A0A091V0J0 A0A2K5NWC4 A0A2K6RDI0 F7HTR8 A0A099ZVY6 A0A0D9RKF6 A0A2K6JQ42 A0A2K5ZYP4 A0A2K5K644 A0A3B3RV35 A0A2K5K656 A0A2K5TLR4 G7MF24 A0A3Q7XQ40 M3YKJ2 A0A384CL93 A0A2K5TLS7 A0A2Y9GGI7 A0A2K6BJH9 F7BH35 G3RN13 A0A2K6BJK2 A0A096NP62 F7BH08 A0A3B4GRR4 A0A1V4JZI6 A0A1V4JZF2 F6TQY1 A0A2I3MCW5 A0A3Q3GU11 A0A3P8PUT5 A0A2K6S2B6 A0A2R8ZPW9 A0A2K6S293 A0A3P9BQE6 W5M0S1 F6UCP4 V8P2G0 A0A452FS37 A0A2I3SUI4 A0A1I7U001 A0A1U7UYH7 A0A3P9NWC9 D3YXG0 F6R9N9 D3YXG0-2 A0A2U3WBH9 A0A1I7U000 A0A2I4B518 S9XZ12 A0A2K5SD05 A0A2Y9JST8 A0A3Q7T881 A0A2K5EMD4 A0A401PF95 L5KZE1 K7G868 W5P565 A0A3B4E5X1 A0A146YJD2 M7BBY2 A0A146YJH4 H0XAK9
A0A2H1VB14 A0A2Z5U737 A0A0N1PJS4 A0A2J7R082 A0A2J7R083 A0A1B6L2U6 A0A2Y9PQD3 A0A341BXZ8 A0A2U4CQ22 A0A2Y9TH90 A0A340WQW6 G1T5K0 Q96RW7 Q96RW7-2 A0A2J8V894 G3QH37 H2Q0S6 K7CJZ0 A0A2J8N6C6 H2N4D8 A0A1B6G7Y9 A0A287DE85 I3MFI4 A0A383YVW7 M3X1M2 F6QWR8 F1PXF9 G1MRA1 A0A2K5NVX0 A0A444UCA5 A0A3Q0CW37 A0A091V0J0 A0A2K5NWC4 A0A2K6RDI0 F7HTR8 A0A099ZVY6 A0A0D9RKF6 A0A2K6JQ42 A0A2K5ZYP4 A0A2K5K644 A0A3B3RV35 A0A2K5K656 A0A2K5TLR4 G7MF24 A0A3Q7XQ40 M3YKJ2 A0A384CL93 A0A2K5TLS7 A0A2Y9GGI7 A0A2K6BJH9 F7BH35 G3RN13 A0A2K6BJK2 A0A096NP62 F7BH08 A0A3B4GRR4 A0A1V4JZI6 A0A1V4JZF2 F6TQY1 A0A2I3MCW5 A0A3Q3GU11 A0A3P8PUT5 A0A2K6S2B6 A0A2R8ZPW9 A0A2K6S293 A0A3P9BQE6 W5M0S1 F6UCP4 V8P2G0 A0A452FS37 A0A2I3SUI4 A0A1I7U001 A0A1U7UYH7 A0A3P9NWC9 D3YXG0 F6R9N9 D3YXG0-2 A0A2U3WBH9 A0A1I7U000 A0A2I4B518 S9XZ12 A0A2K5SD05 A0A2Y9JST8 A0A3Q7T881 A0A2K5EMD4 A0A401PF95 L5KZE1 K7G868 W5P565 A0A3B4E5X1 A0A146YJD2 M7BBY2 A0A146YJH4 H0XAK9
PDB
3DMK
E-value=3.95064e-34,
Score=366
Ontologies
GO
GO:0005509
GO:0007155
GO:0016020
GO:0030054
GO:0009617
GO:0005604
GO:0005938
GO:0032154
GO:0038023
GO:0007156
GO:0005201
GO:0007157
GO:0051301
GO:0007049
GO:0062023
GO:0005913
GO:0050839
GO:0070062
GO:0007601
GO:0042803
GO:0033334
GO:0071711
GO:0016021
GO:0004725
GO:0043589
GO:0090497
GO:0035122
GO:0030198
GO:0005576
GO:0050808
GO:0032589
GO:0005515
GO:0005634
GO:0007067
GO:0006508
GO:0019012
GO:0051603
GO:0030145
GO:0006412
GO:0016876
GO:0043039
PANTHER
Topology
Subcellular location
Secreted
The antibody used to determine subcellular location does not distinguish between HMCN1 and HMCN2. With evidence from 2 publications.
Extracellular space The antibody used to determine subcellular location does not distinguish between HMCN1 and HMCN2. With evidence from 2 publications.
Extracellular matrix The antibody used to determine subcellular location does not distinguish between HMCN1 and HMCN2. With evidence from 2 publications.
Basement membrane The antibody used to determine subcellular location does not distinguish between HMCN1 and HMCN2. With evidence from 2 publications.
Cytoplasm The antibody used to determine subcellular location does not distinguish between HMCN1 and HMCN2. With evidence from 2 publications.
Cell junction The antibody used to determine subcellular location does not distinguish between HMCN1 and HMCN2. With evidence from 2 publications.
Cleavage furrow The antibody used to determine subcellular location does not distinguish between HMCN1 and HMCN2. With evidence from 2 publications.
Extracellular space The antibody used to determine subcellular location does not distinguish between HMCN1 and HMCN2. With evidence from 2 publications.
Extracellular matrix The antibody used to determine subcellular location does not distinguish between HMCN1 and HMCN2. With evidence from 2 publications.
Basement membrane The antibody used to determine subcellular location does not distinguish between HMCN1 and HMCN2. With evidence from 2 publications.
Cytoplasm The antibody used to determine subcellular location does not distinguish between HMCN1 and HMCN2. With evidence from 2 publications.
Cell junction The antibody used to determine subcellular location does not distinguish between HMCN1 and HMCN2. With evidence from 2 publications.
Cleavage furrow The antibody used to determine subcellular location does not distinguish between HMCN1 and HMCN2. With evidence from 2 publications.
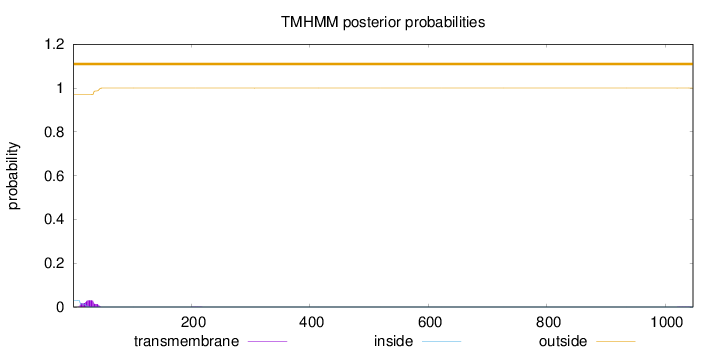
Length:
1047
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.697879999999997
Exp number, first 60 AAs:
0.6868
Total prob of N-in:
0.03038
outside
1 - 1047
Population Genetic Test Statistics
Pi
220.680202
Theta
169.282707
Tajima's D
0.923575
CLR
0.308353
CSRT
0.64216789160542
Interpretation
Uncertain
