Gene
KWMTBOMO08481
Pre Gene Modal
BGIBMGA007650
Annotation
PREDICTED:_piggyBac_transposable_element-derived_protein_3-like_[Papilio_xuthus]
Full name
PiggyBac transposable element-derived protein 3
+ More
Chimeric ERCC6-PGBD3 protein
PiggyBac transposable element-derived protein 2
Chimeric ERCC6-PGBD3 protein
PiggyBac transposable element-derived protein 2
Alternative Name
Chimeric CSB-PGBD3 protein
Location in the cell
Cytoplasmic Reliability : 2.053 Nuclear Reliability : 2.667
Sequence
CDS
ATGGCAAATGACAGACCGTTAGCTGCGCATGAAATTTTAGATGCTTTAGAAAATGTTTCTGATAATGAAGAAGATTATAGAGAACGACTGATATGTATTCTACCTCCTCCTGTTGATCCTGACTGTCTCACTGACGAAGATTCGGGTGAAGAAGATTATGTAACTTTGAATAATTTGCCACGAAACATTCTGCTTCAACCGGCTGAAGTAATGATTCAAGGGCAGATTATGGTGAGTGATACAGAAGAACCTTCTGATTCTACAGATGTTCAAGGCGCTTGCCAAGATAAGCGCCCAATTGAGTGGTTTGAAAACTTCTTAGATGAAGATGTTATTTCGTTGTTGGTGTCAGAGAGCAATAAATATGCTGTCAAAAAGAATTTGCCTGGAGACATAACCACTGAAGATATGAAATGTTTCATCGGCATATTGTTGGTTAGTGGTTATTCATGGCTCCCCCGTAGAATAATGTATTGGGAAAACTCCCCTGATACAAAGAATGAATTGATCAGCTCGGCTATGACTAGGGATAGATTTGACTTTATTTTTCGCCACCTTCATGTCAATGATAATCTGGATTTGCAAGACAAATACACAAAAGTACGCCCCCTAGTTACACTTCTAAATAAAAAGTTCTTAGAGTTTTCTCCTCTTGAAGAGCATTACGGTGTAGATGAGGCCATGATCCCCTACTATGGTAGACATGGCTGCAAACAGCACATAAAAGGTAAACCTATTAGGTACGGGTTCAAAGCTTGGGTTGGTGCTACACGGTTAGGTGAAAAGAGAAACATTGTCATAGAAGAACCACATATGGTGTCCATCTATAACAAATATATGGGAGGAGTGGATCGGTCTGATGAAAATATTTCACATTACCGAATTGGTATACGAGAAAATTTTAGTTTCAGGCTTATCTGGGATATAGACGTGACTGAGTTAAATGAAGTTTTTAATTCACCTGGACATATTGAAGGCTGTTTTTTGTCAAACCCTTTCAGGATTAATGAGTTTAAAATAGCTTTAGAATCTAGAAAAGATAGTACTCCTAGTCTCGATGACATCCCTTAG
Protein
MANDRPLAAHEILDALENVSDNEEDYRERLICILPPPVDPDCLTDEDSGEEDYVTLNNLPRNILLQPAEVMIQGQIMVSDTEEPSDSTDVQGACQDKRPIEWFENFLDEDVISLLVSESNKYAVKKNLPGDITTEDMKCFIGILLVSGYSWLPRRIMYWENSPDTKNELISSAMTRDRFDFIFRHLHVNDNLDLQDKYTKVRPLVTLLNKKFLEFSPLEEHYGVDEAMIPYYGRHGCKQHIKGKPIRYGFKAWVGATRLGEKRNIVIEEPHMVSIYNKYMGGVDRSDENISHYRIGIRENFSFRLIWDIDVTELNEVFNSPGHIEGCFLSNPFRINEFKIALESRKDSTPSLDDIP
Summary
Description
Binds in vitro to PGBD3-related transposable elements, called MER85s; these non-autonomous 140 bp elements are characterized by the presence of PGBD3 terminal inverted repeats and the absence of internal transposase ORF.
Involved in repair of DNA damage following UV irradiation, acting either in the absence of ERCC6 or synergistically with ERCC6. Involved in the regulation of gene expression. In the absence of ERCC6, induces the expression of genes characteristic of interferon-like antiviral responses. This response is almost completely suppressed in the presence of ERCC6. In the presence of ERCC6, regulates the expression of genes involved in metabolism regulation, including IGFBP5 and IGFBP7. In vitro binds to PGBD3-related transposable elements, called MER85s; these non-autonomous 140 bp elements are characterized by the presence of PGBD3 terminal inverted repeats and the absence of internal transposase ORF.
Involved in repair of DNA damage following UV irradiation, acting either in the absence of ERCC6 or synergistically with ERCC6. Involved in the regulation of gene expression. In the absence of ERCC6, induces the expression of genes characteristic of interferon-like antiviral responses. This response is almost completely suppressed in the presence of ERCC6. In the presence of ERCC6, regulates the expression of genes involved in metabolism regulation, including IGFBP5 and IGFBP7. In vitro binds to PGBD3-related transposable elements, called MER85s; these non-autonomous 140 bp elements are characterized by the presence of PGBD3 terminal inverted repeats and the absence of internal transposase ORF.
Miscellaneous
PGBD3 gene is located within ERCC6 intron 5.
Keywords
Alternative splicing
Complete proteome
DNA-binding
Nucleus
Phosphoprotein
Polymorphism
Reference proteome
Disease mutation
Isopeptide bond
Premature ovarian failure
Ubl conjugation
Feature
chain PiggyBac transposable element-derived protein 3
sequence variant In dbSNP:rs4253072.
splice variant In isoform 2.
sequence variant In dbSNP:rs4253072.
splice variant In isoform 2.
Uniprot
A0A3S2N5S4
A0A212FJ76
A0A1A9Z8F7
A0A1Y1LG12
T2M9K1
F7EM96
+ More
A0A2J8JGQ8 Q8N328 U3B6D7 U3E686 U3ENV5 U3CPS9 A0A2K6E0X9 A0A2J8RT17 K7C897 A0A2R8Z9V9 A0A2K5QFQ3 A0A147BBG9 G3SEC0 A0A2K6E0V7 P0DP91 A8K4Q3 A0A2K5V985 A0A2I3RAX8 A0A2J8JGN8 A0A2K5DEG0 A0A2K5JRR6 A0A2K6PI71 A0A2K6LGX8 F6RSP5 A0A2K6ST22 Q4R4A3 A0A2J8RT31 A0A096P4A2 A0A2K6AE02 A0A2I3HPQ9 D7EK33 A0A2K5LL76 N6T2W4 A0A087U8G4 F6ZNL0 A0A336M992 M3XQ58 A0A3P4RWH6 M3X8R6 G3VIF3 A0A2Y9MDI9 A0A2Y9MJ53 S9WNB2 A0A212ERY7 A0A2Y9S0G5 A0A2U4BIF0 A0A2U4BI88 A0A452DQQ1 A0A341BUQ3 A0A341BW94 G5E5N2 A0A2K6F4D2 F7I1C5 L8IPN2 A0A2K6UA69 A0A212D1C8 A0A2Y9HA29 A0A340X956 A0A2U3W5X2 A0A2Y9IGG0 W5PEK4 A0A2K5KAR4 A0A383Z4K2 K9J215 A0A3Q7MPJ8 A0A2K5Q526 A0A2U3XGG7 A0A2K5Q520 A0A3Q2HLG4 F6X578 G3SS70 B4DPA1 A0A3Q7RRM6 A0A2K6KZJ0 G1QWB6 Q6P3X8 H2N335 A0A2J8RJL9 I3N029 K7AM28 A0A0D9R971 H2Q1I5 G3RPE5 D2I5Z2 A0A2K6A111
A0A2J8JGQ8 Q8N328 U3B6D7 U3E686 U3ENV5 U3CPS9 A0A2K6E0X9 A0A2J8RT17 K7C897 A0A2R8Z9V9 A0A2K5QFQ3 A0A147BBG9 G3SEC0 A0A2K6E0V7 P0DP91 A8K4Q3 A0A2K5V985 A0A2I3RAX8 A0A2J8JGN8 A0A2K5DEG0 A0A2K5JRR6 A0A2K6PI71 A0A2K6LGX8 F6RSP5 A0A2K6ST22 Q4R4A3 A0A2J8RT31 A0A096P4A2 A0A2K6AE02 A0A2I3HPQ9 D7EK33 A0A2K5LL76 N6T2W4 A0A087U8G4 F6ZNL0 A0A336M992 M3XQ58 A0A3P4RWH6 M3X8R6 G3VIF3 A0A2Y9MDI9 A0A2Y9MJ53 S9WNB2 A0A212ERY7 A0A2Y9S0G5 A0A2U4BIF0 A0A2U4BI88 A0A452DQQ1 A0A341BUQ3 A0A341BW94 G5E5N2 A0A2K6F4D2 F7I1C5 L8IPN2 A0A2K6UA69 A0A212D1C8 A0A2Y9HA29 A0A340X956 A0A2U3W5X2 A0A2Y9IGG0 W5PEK4 A0A2K5KAR4 A0A383Z4K2 K9J215 A0A3Q7MPJ8 A0A2K5Q526 A0A2U3XGG7 A0A2K5Q520 A0A3Q2HLG4 F6X578 G3SS70 B4DPA1 A0A3Q7RRM6 A0A2K6KZJ0 G1QWB6 Q6P3X8 H2N335 A0A2J8RJL9 I3N029 K7AM28 A0A0D9R971 H2Q1I5 G3RPE5 D2I5Z2 A0A2K6A111
Pubmed
22118469
28004739
24065732
17431167
25319552
14702039
+ More
15164054 15489334 18369450 22483866 23186163 26218421 16959974 25243066 22722832 29652888 22398555 17081983 16964243 18691976 18669648 20068231 21406692 28112733 11181995 16136131 25362486 15944441 18362917 19820115 23537049 17495919 17975172 21709235 23149746 19393038 22751099 20809919 19892987 17974005 16710414 20010809
15164054 15489334 18369450 22483866 23186163 26218421 16959974 25243066 22722832 29652888 22398555 17081983 16964243 18691976 18669648 20068231 21406692 28112733 11181995 16136131 25362486 15944441 18362917 19820115 23537049 17495919 17975172 21709235 23149746 19393038 22751099 20809919 19892987 17974005 16710414 20010809
EMBL
RSAL01000353
RVE42252.1
AGBW02008302
OWR53786.1
GEZM01060808
GEZM01060807
+ More
JAV70895.1 HAAD01002582 CDG68814.1 JSUE03043349 JU470545 AFH27349.1 NBAG03000461 PNI21937.1 AK074682 AL138760 BC028954 BC063690 GAMT01003553 GAMT01003551 GAMQ01002282 JAB08308.1 JAB39569.1 GAMS01002010 JAB21126.1 GAMP01000187 JAB52568.1 GAMR01006104 JAB27828.1 NDHI03003654 PNJ11666.1 GABF01001690 JAA20455.1 GEGO01007284 JAR88120.1 CABD030070918 BC034479 AK291018 CH471187 BAF83707.1 EAW93095.1 AQIA01070964 AACZ04060518 PNI21935.1 PNI21936.1 AB179012 BAE02063.1 PNJ11664.1 PNJ11665.1 AHZZ02035229 ADFV01105741 ADFV01105742 ADFV01105743 ADFV01105744 ADFV01105745 DS497727 EFA12977.1 APGK01046441 KB741054 KB632013 ENN74459.1 ERL88014.1 KK118708 KFM73653.1 UFQT01000583 SSX25489.1 AEYP01099555 AEYP01099556 CYRY02043934 VCX38622.1 AANG04001781 AEFK01164926 AEFK01164927 KB017432 EPY77693.1 AGBW02012897 OWR44258.1 LWLT01000008 GAMT01004683 GAMP01007477 JAB07178.1 JAB45278.1 JH880825 ELR58540.1 MKHE01000009 OWK12058.1 AMGL01092670 GABZ01005161 JAA48364.1 AK298248 BAG60513.1 ADFV01037738 ADFV01037739 ADFV01037740 ADFV01037741 ADFV01037742 AK123219 BX647065 AL672183 CH471257 BC063785 ABGA01061445 ABGA01061446 ABGA01061447 NDHI03003683 PNJ08718.1 AGTP01101535 AGTP01101536 AACZ04070590 GABC01008499 GABE01003522 JAA02839.1 JAA41217.1 AQIB01155464 CABD030010211 CABD030010212 CABD030010213 GL194834 EFB16732.1
JAV70895.1 HAAD01002582 CDG68814.1 JSUE03043349 JU470545 AFH27349.1 NBAG03000461 PNI21937.1 AK074682 AL138760 BC028954 BC063690 GAMT01003553 GAMT01003551 GAMQ01002282 JAB08308.1 JAB39569.1 GAMS01002010 JAB21126.1 GAMP01000187 JAB52568.1 GAMR01006104 JAB27828.1 NDHI03003654 PNJ11666.1 GABF01001690 JAA20455.1 GEGO01007284 JAR88120.1 CABD030070918 BC034479 AK291018 CH471187 BAF83707.1 EAW93095.1 AQIA01070964 AACZ04060518 PNI21935.1 PNI21936.1 AB179012 BAE02063.1 PNJ11664.1 PNJ11665.1 AHZZ02035229 ADFV01105741 ADFV01105742 ADFV01105743 ADFV01105744 ADFV01105745 DS497727 EFA12977.1 APGK01046441 KB741054 KB632013 ENN74459.1 ERL88014.1 KK118708 KFM73653.1 UFQT01000583 SSX25489.1 AEYP01099555 AEYP01099556 CYRY02043934 VCX38622.1 AANG04001781 AEFK01164926 AEFK01164927 KB017432 EPY77693.1 AGBW02012897 OWR44258.1 LWLT01000008 GAMT01004683 GAMP01007477 JAB07178.1 JAB45278.1 JH880825 ELR58540.1 MKHE01000009 OWK12058.1 AMGL01092670 GABZ01005161 JAA48364.1 AK298248 BAG60513.1 ADFV01037738 ADFV01037739 ADFV01037740 ADFV01037741 ADFV01037742 AK123219 BX647065 AL672183 CH471257 BC063785 ABGA01061445 ABGA01061446 ABGA01061447 NDHI03003683 PNJ08718.1 AGTP01101535 AGTP01101536 AACZ04070590 GABC01008499 GABE01003522 JAA02839.1 JAA41217.1 AQIB01155464 CABD030010211 CABD030010212 CABD030010213 GL194834 EFB16732.1
Proteomes
UP000283053
UP000007151
UP000092445
UP000006718
UP000005640
UP000233120
+ More
UP000240080 UP000233040 UP000001519 UP000233100 UP000002277 UP000233020 UP000233080 UP000233200 UP000233180 UP000008225 UP000233220 UP000028761 UP000233140 UP000001073 UP000007266 UP000233060 UP000019118 UP000030742 UP000054359 UP000002280 UP000000715 UP000011712 UP000007648 UP000248483 UP000248484 UP000245320 UP000291000 UP000252040 UP000009136 UP000233160 UP000248481 UP000265300 UP000245340 UP000248482 UP000002356 UP000261681 UP000286641 UP000245341 UP000002281 UP000007646 UP000286640 UP000001595 UP000005215 UP000029965
UP000240080 UP000233040 UP000001519 UP000233100 UP000002277 UP000233020 UP000233080 UP000233200 UP000233180 UP000008225 UP000233220 UP000028761 UP000233140 UP000001073 UP000007266 UP000233060 UP000019118 UP000030742 UP000054359 UP000002280 UP000000715 UP000011712 UP000007648 UP000248483 UP000248484 UP000245320 UP000291000 UP000252040 UP000009136 UP000233160 UP000248481 UP000265300 UP000245340 UP000248482 UP000002356 UP000261681 UP000286641 UP000245341 UP000002281 UP000007646 UP000286640 UP000001595 UP000005215 UP000029965
Pfam
PF13843 DDE_Tnp_1_7
Interpro
IPR029526
PGBD
ProteinModelPortal
A0A3S2N5S4
A0A212FJ76
A0A1A9Z8F7
A0A1Y1LG12
T2M9K1
F7EM96
+ More
A0A2J8JGQ8 Q8N328 U3B6D7 U3E686 U3ENV5 U3CPS9 A0A2K6E0X9 A0A2J8RT17 K7C897 A0A2R8Z9V9 A0A2K5QFQ3 A0A147BBG9 G3SEC0 A0A2K6E0V7 P0DP91 A8K4Q3 A0A2K5V985 A0A2I3RAX8 A0A2J8JGN8 A0A2K5DEG0 A0A2K5JRR6 A0A2K6PI71 A0A2K6LGX8 F6RSP5 A0A2K6ST22 Q4R4A3 A0A2J8RT31 A0A096P4A2 A0A2K6AE02 A0A2I3HPQ9 D7EK33 A0A2K5LL76 N6T2W4 A0A087U8G4 F6ZNL0 A0A336M992 M3XQ58 A0A3P4RWH6 M3X8R6 G3VIF3 A0A2Y9MDI9 A0A2Y9MJ53 S9WNB2 A0A212ERY7 A0A2Y9S0G5 A0A2U4BIF0 A0A2U4BI88 A0A452DQQ1 A0A341BUQ3 A0A341BW94 G5E5N2 A0A2K6F4D2 F7I1C5 L8IPN2 A0A2K6UA69 A0A212D1C8 A0A2Y9HA29 A0A340X956 A0A2U3W5X2 A0A2Y9IGG0 W5PEK4 A0A2K5KAR4 A0A383Z4K2 K9J215 A0A3Q7MPJ8 A0A2K5Q526 A0A2U3XGG7 A0A2K5Q520 A0A3Q2HLG4 F6X578 G3SS70 B4DPA1 A0A3Q7RRM6 A0A2K6KZJ0 G1QWB6 Q6P3X8 H2N335 A0A2J8RJL9 I3N029 K7AM28 A0A0D9R971 H2Q1I5 G3RPE5 D2I5Z2 A0A2K6A111
A0A2J8JGQ8 Q8N328 U3B6D7 U3E686 U3ENV5 U3CPS9 A0A2K6E0X9 A0A2J8RT17 K7C897 A0A2R8Z9V9 A0A2K5QFQ3 A0A147BBG9 G3SEC0 A0A2K6E0V7 P0DP91 A8K4Q3 A0A2K5V985 A0A2I3RAX8 A0A2J8JGN8 A0A2K5DEG0 A0A2K5JRR6 A0A2K6PI71 A0A2K6LGX8 F6RSP5 A0A2K6ST22 Q4R4A3 A0A2J8RT31 A0A096P4A2 A0A2K6AE02 A0A2I3HPQ9 D7EK33 A0A2K5LL76 N6T2W4 A0A087U8G4 F6ZNL0 A0A336M992 M3XQ58 A0A3P4RWH6 M3X8R6 G3VIF3 A0A2Y9MDI9 A0A2Y9MJ53 S9WNB2 A0A212ERY7 A0A2Y9S0G5 A0A2U4BIF0 A0A2U4BI88 A0A452DQQ1 A0A341BUQ3 A0A341BW94 G5E5N2 A0A2K6F4D2 F7I1C5 L8IPN2 A0A2K6UA69 A0A212D1C8 A0A2Y9HA29 A0A340X956 A0A2U3W5X2 A0A2Y9IGG0 W5PEK4 A0A2K5KAR4 A0A383Z4K2 K9J215 A0A3Q7MPJ8 A0A2K5Q526 A0A2U3XGG7 A0A2K5Q520 A0A3Q2HLG4 F6X578 G3SS70 B4DPA1 A0A3Q7RRM6 A0A2K6KZJ0 G1QWB6 Q6P3X8 H2N335 A0A2J8RJL9 I3N029 K7AM28 A0A0D9R971 H2Q1I5 G3RPE5 D2I5Z2 A0A2K6A111
Ontologies
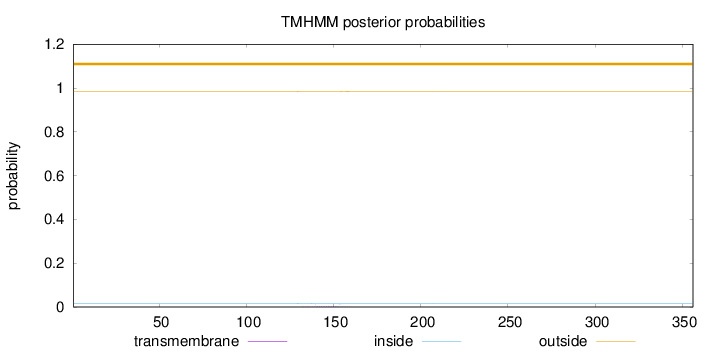
Topology
Subcellular location
Nucleus
Length:
356
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02877
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01543
outside
1 - 356
Population Genetic Test Statistics
Pi
395.177149
Theta
200.316456
Tajima's D
3.029588
CLR
0.380344
CSRT
0.978501074946253
Interpretation
Possibly Balancing Selection
