Gene
KWMTBOMO08477
Annotation
polymerase_[Lishi_Spider_Virus_1]
Full name
Large structural protein
+ More
RNA-directed RNA polymerase L
RNA-directed RNA polymerase L
Alternative Name
RNA-directed RNA polymerase L
Large structural protein
Replicase
Transcriptase
Large structural protein
Replicase
Transcriptase
Location in the cell
Mitochondrial Reliability : 1.786 Nuclear Reliability : 1.758
Sequence
CDS
ATGCCAGCGTTACCGTCTACAGTGTTGAGAAATAAGATGGAAACATTATTGCCTAAAATCACTAAGAATCAAGACGTCAAGAATCTGATTCAGGCTGCTAAATCAGTGTCCAGTCAGGAGTTCATTAAGGCCTTAGATTCATCTAATATATTACCTGCTCGTGTATTTGCTGCTCTATATGCAGCAAGTCCTAAAGGGATGTTGTCGGAATTTCTCGCAAAGTTTGAAACAGCAAGAAGTGTGTTACAAGCACTTATCTTAGGAAGCGGAAAGTATGCGGCTAGAAAAATGCTAAGACGACTAGTTAAACAGGAACACATTTTACAGAACTGGAGAGTCAATGTTATAAAAGGTCATGTTCGTGGTTACGATTATATGCATATTATCCTAGGTAACAAGTGCCCAGGAAAGGCCGCGAACTTAATTAGGGAAGAGTCATGGCGAAAACCTGTTGAGACAGTCACTATGCCTCCTCTTCAGCACCAAGTATATATGACAACACCTGAAAGAGGATCATTGAGTATGCATGATATCAAAAGTCATTTTACAATTAAATGTAACCAGTCCACAACTACCATTACCGAAGACGGGCGTCCTCACTACAGCACAGGGAATTTTAGACCTTTCGAGGGATACACTACCAGAACTGGTACCGTGGAGCCCACTATGAGTTTTATTGAGAAGGATGTTTTACTATCGAAACTGCAAAACGTCATAGAATTGGCTTCATGGACAAACAAGACTGGACTGAACAAAGAGAAGACTGCATCAATCAACAGTAATCTGCATCTGCTGATAGCTAAAATTGTTGAACAACACACTGATCATGACTTGAAGGTATTTGCACCATTCGTTGGCTGCCGACGATCTGGTACAGTTCAACATCATATCAGATGTCCAGGATACAGAGAGTCTATAGTCCCCAATACTCTACTAAATATGTATACACATACAGAGGGAGAGTCTGATTCCAACATGCATGTGAACTCCCAGCCTGGACAACATTACAAGATTAATTTCCTACACATCTACTGTTATGCTGTAGTTAGCGTCTTCTTGCACAATGAGATTTATTGTACGCCATCAGACAATAAAATCGTATGGTGTGTAACCCGGGATTGTGAGGAGTGTATGAGCCCTATTGATAAGACCCCGATTGTCATAGATGAGTCACTAATTGCAGATATAGATTTCTCGGCATTGAAGATCACTACATTAACAGAAGATGCGATGGATGTACTCAGGAAATCATTGGATATTCATGAAGGACGTGTCTATATGGTACCTGATGAAGAGACTAGGCTAGAAAATCAAATTGCATGTGAGGGAGTAATCCAAGAATTGTTGGAGACATTAGTGATGTCTAAAAATGCCTTGATTACACGATACACTCAGCATGCAATGACAAATGAAGGAAAACAAGTGTTAGGGGCGTTTGCAGTGAAGAATAAAAGGAGAGAGATTGGATTATCGGAATAA
Protein
MPALPSTVLRNKMETLLPKITKNQDVKNLIQAAKSVSSQEFIKALDSSNILPARVFAALYAASPKGMLSEFLAKFETARSVLQALILGSGKYAARKMLRRLVKQEHILQNWRVNVIKGHVRGYDYMHIILGNKCPGKAANLIREESWRKPVETVTMPPLQHQVYMTTPERGSLSMHDIKSHFTIKCNQSTTTITEDGRPHYSTGNFRPFEGYTTRTGTVEPTMSFIEKDVLLSKLQNVIELASWTNKTGLNKEKTASINSNLHLLIAKIVEQHTDHDLKVFAPFVGCRRSGTVQHHIRCPGYRESIVPNTLLNMYTHTEGESDSNMHVNSQPGQHYKINFLHIYCYAVVSVFLHNEIYCTPSDNKIVWCVTRDCEECMSPIDKTPIVIDESLIADIDFSALKITTLTEDAMDVLRKSLDIHEGRVYMVPDEETRLENQIACEGVIQELLETLVMSKNALITRYTQHAMTNEGKQVLGAFAVKNKRREIGLSE
Summary
Description
RNA-directed RNA polymerase that catalyzes the transcription of viral mRNAs, their capping and polyadenylation. The template is composed of the viral RNA tightly encapsidated by the nucleoprotein (N). The viral polymerase binds to the genomic RNA at the 3' leader promoter, and transcribes subsequently all viral mRNAs with a decreasing efficiency. The first gene is the most transcribed, and the last the least transcribed. The viral phosphoprotein acts as a processivity factor. Capping is concommitant with initiation of mRNA transcription. Indeed, a GDP polyribonucleotidyl transferase (PRNTase) adds the cap structure when the nascent RNA chain length has reached few nucleotides. Ribose 2'-O methylation of viral mRNA cap precedes and facilitates subsequent guanine-N-7 methylation, both activities being carried by the viral polymerase. Polyadenylation of mRNAs occur by a stuttering mechanism at a slipery stop site present at the end viral genes. After finishing transcription of a mRNA, the polymerase can resume transcription of the downstream gene.
Catalytic Activity
a ribonucleoside 5'-triphosphate + RNA(n) = diphosphate + RNA(n+1)
a 5'-(5'-triphosphoguanosine)-(purine-ribonucleotide)-[mRNA] + S-adenosyl-L-methionine = a 5'-(N(7)-methyl 5'-triphosphoguanosine)-(purine-ribonucleotide)-[mRNA] + S-adenosyl-L-homocysteine
a 5'-(N(7)-methyl 5'-triphosphoguanosine)-(2'-O-methyl-purine-ribonucleotide)-(ribonucleotide)-[mRNA] + S-adenosyl-L-methionine = a 5'-(N(7)-methyl 5'-triphosphoguanosine)-(2'-O-methyl-purine-ribonucleotide)-(2'-O-methyl-ribonucleotide)-[mRNA] + H(+) + S-adenosyl-L-homocysteine
a 5'-triphospho-(purine-ribonucleotide)-[mRNA] + GDP + H(+) = a 5'-(5'-triphosphoguanosine)-(purine-ribonucleotide)-[mRNA] + diphosphate
a 5'-(5'-triphosphoguanosine)-(purine-ribonucleotide)-[mRNA] + S-adenosyl-L-methionine = a 5'-(N(7)-methyl 5'-triphosphoguanosine)-(purine-ribonucleotide)-[mRNA] + S-adenosyl-L-homocysteine
a 5'-(N(7)-methyl 5'-triphosphoguanosine)-(2'-O-methyl-purine-ribonucleotide)-(ribonucleotide)-[mRNA] + S-adenosyl-L-methionine = a 5'-(N(7)-methyl 5'-triphosphoguanosine)-(2'-O-methyl-purine-ribonucleotide)-(2'-O-methyl-ribonucleotide)-[mRNA] + H(+) + S-adenosyl-L-homocysteine
a 5'-triphospho-(purine-ribonucleotide)-[mRNA] + GDP + H(+) = a 5'-(5'-triphosphoguanosine)-(purine-ribonucleotide)-[mRNA] + diphosphate
Similarity
Belongs to the paramyxovirus L protein family.
Uniprot
A0A1L3KMW6
A0A0B5KRA0
A0A1L4A1T3
A0A2K9YNF4
A0A2S0SZ96
A0A0B5KTA1
+ More
A0A1L3KN70 A0A1L3KMX9 A0A1L3KN74 A0A221LFF1 A0A0B5KEQ8 A0A0S2RRG3 A0A1L3KN44 A0A346M252 A0A346M266 A0A0B5KJW3 A0A221LFF6 A0A2P1GMT0 A0A2P1GMQ1 A0A1L3KMY9 A0A2P1GMQ7 A0A0B5KX86 A0A1L3KMQ9 A0A0B5KT91 A0A0B5KT83 A0A0B5KEM5 A0A3G3BTF8 A0A1L3KN64 A0A385DMP2 A0A2U8JHB6 A0A0B5KR88 A0A076E9Z7 A0A0A1G6U7 A0A075EJ04 A0A0B5KX99 A0A3G4YIX7 A0A3G4YJ38 A0A3G4YJ66 A0A0B5KTB0 A0A172MHM9 A0A3G3BTR3 A0A0B5KJX3 A0A3G4YJ20 A0A1L3KN56 A0A1L3KMX3 A0A1L3KN37 A0A1L3KN90 A0A3G5FMI8 A0A1L3KMV0 A0A3B1EJ88 A0A1L3KML0 A0A2S1ZPS1 C4NFK9 C4NFL5 A0A2Z4HF79 A0A2Z4HFC4 A0A2Z4HFJ7 A0A2Z4HF97 A0A2Z4HF69 A0A2Z4HFG4 A0A2Z4GIN2 A0A2Z4HFS7 A0A2H4YI41 A0A2Z4HFJ1 A0A2Z4HFC6 A0A0B5KEN7 A0A2Z4HFA6 A0A2Z4HFC7 A0A2Z4HFB8 A0A2Z4HFG9 A0A1L3KMU5 A0A0B5KRF3 A0A060DA11 A0A1L3KN23 A0A0X3PRB1 A0A067YEZ6 A0A1Z2RTE0 A0A0X3PYN5 A0A1Z2RSU8 A0A1Z2RTB4 A0A1Z2RT54 A0A1Z2RT41 A0A2Z4Z3N4 A0A0B5KTH4 A0A1B1FIU1 A0A0B5KK67
A0A1L3KN70 A0A1L3KMX9 A0A1L3KN74 A0A221LFF1 A0A0B5KEQ8 A0A0S2RRG3 A0A1L3KN44 A0A346M252 A0A346M266 A0A0B5KJW3 A0A221LFF6 A0A2P1GMT0 A0A2P1GMQ1 A0A1L3KMY9 A0A2P1GMQ7 A0A0B5KX86 A0A1L3KMQ9 A0A0B5KT91 A0A0B5KT83 A0A0B5KEM5 A0A3G3BTF8 A0A1L3KN64 A0A385DMP2 A0A2U8JHB6 A0A0B5KR88 A0A076E9Z7 A0A0A1G6U7 A0A075EJ04 A0A0B5KX99 A0A3G4YIX7 A0A3G4YJ38 A0A3G4YJ66 A0A0B5KTB0 A0A172MHM9 A0A3G3BTR3 A0A0B5KJX3 A0A3G4YJ20 A0A1L3KN56 A0A1L3KMX3 A0A1L3KN37 A0A1L3KN90 A0A3G5FMI8 A0A1L3KMV0 A0A3B1EJ88 A0A1L3KML0 A0A2S1ZPS1 C4NFK9 C4NFL5 A0A2Z4HF79 A0A2Z4HFC4 A0A2Z4HFJ7 A0A2Z4HF97 A0A2Z4HF69 A0A2Z4HFG4 A0A2Z4GIN2 A0A2Z4HFS7 A0A2H4YI41 A0A2Z4HFJ1 A0A2Z4HFC6 A0A0B5KEN7 A0A2Z4HFA6 A0A2Z4HFC7 A0A2Z4HFB8 A0A2Z4HFG9 A0A1L3KMU5 A0A0B5KRF3 A0A060DA11 A0A1L3KN23 A0A0X3PRB1 A0A067YEZ6 A0A1Z2RTE0 A0A0X3PYN5 A0A1Z2RSU8 A0A1Z2RTB4 A0A1Z2RT54 A0A1Z2RT41 A0A2Z4Z3N4 A0A0B5KTH4 A0A1B1FIU1 A0A0B5KK67
Pubmed
EMBL
KX884427
APG78724.1
KM817597
AJG39051.1
KX924630
API61887.1
+ More
MF360789 AUW34382.1 MF416404 AWB14664.1 KM817604 AJG39067.1 KX884451 APG78824.1 KX884424 APG78716.1 KX884453 APG78828.1 MF189989 ASM94010.1 KM817610 AJG39074.1 KU095839 ALP32028.1 KX884455 APG78831.1 MH188031 AXQ04827.1 MH188036 AXQ04841.1 KM817601 AJG39060.1 MF189987 ASM94008.1 MG600011 AVM87278.1 MG600010 AVM87275.1 KX884439 APG78770.1 MG600009 AVM87272.1 KM817600 AJG39058.1 KX884419 APG78700.1 KM817599 AJG39057.1 KM817595 AJG39047.1 KM817594 AJG39044.1 MK026591 AYP67566.1 KX884449 APG78818.1 MG880117 AXQ59273.1 MG764515 AWK68105.1 KM817593 MH688555 AJG39041.1 QBQ65118.1 KM048317 AII01805.1 KM460042 AIY53910.1 KJ746903 AIE42676.2 KM817609 AJG39073.1 MH155923 AYV61054.1 MH155927 AYV61060.1 MH155920 AYV61047.1 KM817611 AJG39077.1 KU230451 ANC97697.1 MK026566 AYP67535.1 KM817606 AJG39070.1 MH155921 AYV61050.1 KX884458 APG78840.1 KX884409 APG78655.1 KX884444 APG78798.1 KX884461 APG78852.1 MH477287 AYW51538.1 KX884404 APG78635.1 MF287670 AWI42881.1 KX884408 APG78650.1 MH237595 AWK27462.1 FJ554525 ACQ94979.1 FJ554526 ACQ94985.1 MH430658 AWW13489.1 MH430666 AWW13507.1 MH430650 AWW13450.1 MH430653 AWW13465.1 MH430655 AWW13479.1 MH430648 MH430659 AWW13443.1 AWW13492.1 MH037149 AWW01093.1 MH430652 AWW13458.1 MG012486 AUE23903.1 MH430665 AWW13503.1 MH430657 AWW13485.1 KM817598 AJG39054.1 MH430663 MH430664 AWW13497.1 AWW13500.1 MH430656 AWW13482.1 MH430654 AWW13472.1 MH430651 AWW13453.1 KX884418 APG78697.1 KM817632 AJG39111.1 KF823814 AIB06812.1 KX884441 APG78782.1 GEEE01008921 JAP54304.1 KF530058 AHA90827.1 MF176375 ASA47494.1 GEEE01006270 JAP56955.1 MF176245 ASA47282.1 MF176356 ASA47464.1 MF176316 ASA47403.1 MF176296 ASA47369.1 MH213246 AXA52562.1 KM817641 AJG39142.1 KX257488 ANQ45640.1 KM817645 AJG39155.1
MF360789 AUW34382.1 MF416404 AWB14664.1 KM817604 AJG39067.1 KX884451 APG78824.1 KX884424 APG78716.1 KX884453 APG78828.1 MF189989 ASM94010.1 KM817610 AJG39074.1 KU095839 ALP32028.1 KX884455 APG78831.1 MH188031 AXQ04827.1 MH188036 AXQ04841.1 KM817601 AJG39060.1 MF189987 ASM94008.1 MG600011 AVM87278.1 MG600010 AVM87275.1 KX884439 APG78770.1 MG600009 AVM87272.1 KM817600 AJG39058.1 KX884419 APG78700.1 KM817599 AJG39057.1 KM817595 AJG39047.1 KM817594 AJG39044.1 MK026591 AYP67566.1 KX884449 APG78818.1 MG880117 AXQ59273.1 MG764515 AWK68105.1 KM817593 MH688555 AJG39041.1 QBQ65118.1 KM048317 AII01805.1 KM460042 AIY53910.1 KJ746903 AIE42676.2 KM817609 AJG39073.1 MH155923 AYV61054.1 MH155927 AYV61060.1 MH155920 AYV61047.1 KM817611 AJG39077.1 KU230451 ANC97697.1 MK026566 AYP67535.1 KM817606 AJG39070.1 MH155921 AYV61050.1 KX884458 APG78840.1 KX884409 APG78655.1 KX884444 APG78798.1 KX884461 APG78852.1 MH477287 AYW51538.1 KX884404 APG78635.1 MF287670 AWI42881.1 KX884408 APG78650.1 MH237595 AWK27462.1 FJ554525 ACQ94979.1 FJ554526 ACQ94985.1 MH430658 AWW13489.1 MH430666 AWW13507.1 MH430650 AWW13450.1 MH430653 AWW13465.1 MH430655 AWW13479.1 MH430648 MH430659 AWW13443.1 AWW13492.1 MH037149 AWW01093.1 MH430652 AWW13458.1 MG012486 AUE23903.1 MH430665 AWW13503.1 MH430657 AWW13485.1 KM817598 AJG39054.1 MH430663 MH430664 AWW13497.1 AWW13500.1 MH430656 AWW13482.1 MH430654 AWW13472.1 MH430651 AWW13453.1 KX884418 APG78697.1 KM817632 AJG39111.1 KF823814 AIB06812.1 KX884441 APG78782.1 GEEE01008921 JAP54304.1 KF530058 AHA90827.1 MF176375 ASA47494.1 GEEE01006270 JAP56955.1 MF176245 ASA47282.1 MF176356 ASA47464.1 MF176316 ASA47403.1 MF176296 ASA47369.1 MH213246 AXA52562.1 KM817641 AJG39142.1 KX257488 ANQ45640.1 KM817645 AJG39155.1
Proteomes
UP000202515
UP000279202
UP000272305
UP000204621
UP000202207
UP000203961
+ More
UP000204649 UP000201479 UP000203655 UP000280558 UP000202294 UP000201991 UP000202577 UP000207671 UP000202602 UP000214367 UP000204161 UP000204131 UP000218499 UP000276326 UP000202848 UP000277466 UP000201870 UP000204661 UP000201961 UP000203174 UP000203332 UP000279435 UP000119000 UP000029765 UP000201788 UP000204289 UP000232260 UP000113864 UP000202564 UP000127615 UP000204249 UP000278361
UP000204649 UP000201479 UP000203655 UP000280558 UP000202294 UP000201991 UP000202577 UP000207671 UP000202602 UP000214367 UP000204161 UP000204131 UP000218499 UP000276326 UP000202848 UP000277466 UP000201870 UP000204661 UP000201961 UP000203174 UP000203332 UP000279435 UP000119000 UP000029765 UP000201788 UP000204289 UP000232260 UP000113864 UP000202564 UP000127615 UP000204249 UP000278361
Interpro
ProteinModelPortal
A0A1L3KMW6
A0A0B5KRA0
A0A1L4A1T3
A0A2K9YNF4
A0A2S0SZ96
A0A0B5KTA1
+ More
A0A1L3KN70 A0A1L3KMX9 A0A1L3KN74 A0A221LFF1 A0A0B5KEQ8 A0A0S2RRG3 A0A1L3KN44 A0A346M252 A0A346M266 A0A0B5KJW3 A0A221LFF6 A0A2P1GMT0 A0A2P1GMQ1 A0A1L3KMY9 A0A2P1GMQ7 A0A0B5KX86 A0A1L3KMQ9 A0A0B5KT91 A0A0B5KT83 A0A0B5KEM5 A0A3G3BTF8 A0A1L3KN64 A0A385DMP2 A0A2U8JHB6 A0A0B5KR88 A0A076E9Z7 A0A0A1G6U7 A0A075EJ04 A0A0B5KX99 A0A3G4YIX7 A0A3G4YJ38 A0A3G4YJ66 A0A0B5KTB0 A0A172MHM9 A0A3G3BTR3 A0A0B5KJX3 A0A3G4YJ20 A0A1L3KN56 A0A1L3KMX3 A0A1L3KN37 A0A1L3KN90 A0A3G5FMI8 A0A1L3KMV0 A0A3B1EJ88 A0A1L3KML0 A0A2S1ZPS1 C4NFK9 C4NFL5 A0A2Z4HF79 A0A2Z4HFC4 A0A2Z4HFJ7 A0A2Z4HF97 A0A2Z4HF69 A0A2Z4HFG4 A0A2Z4GIN2 A0A2Z4HFS7 A0A2H4YI41 A0A2Z4HFJ1 A0A2Z4HFC6 A0A0B5KEN7 A0A2Z4HFA6 A0A2Z4HFC7 A0A2Z4HFB8 A0A2Z4HFG9 A0A1L3KMU5 A0A0B5KRF3 A0A060DA11 A0A1L3KN23 A0A0X3PRB1 A0A067YEZ6 A0A1Z2RTE0 A0A0X3PYN5 A0A1Z2RSU8 A0A1Z2RTB4 A0A1Z2RT54 A0A1Z2RT41 A0A2Z4Z3N4 A0A0B5KTH4 A0A1B1FIU1 A0A0B5KK67
A0A1L3KN70 A0A1L3KMX9 A0A1L3KN74 A0A221LFF1 A0A0B5KEQ8 A0A0S2RRG3 A0A1L3KN44 A0A346M252 A0A346M266 A0A0B5KJW3 A0A221LFF6 A0A2P1GMT0 A0A2P1GMQ1 A0A1L3KMY9 A0A2P1GMQ7 A0A0B5KX86 A0A1L3KMQ9 A0A0B5KT91 A0A0B5KT83 A0A0B5KEM5 A0A3G3BTF8 A0A1L3KN64 A0A385DMP2 A0A2U8JHB6 A0A0B5KR88 A0A076E9Z7 A0A0A1G6U7 A0A075EJ04 A0A0B5KX99 A0A3G4YIX7 A0A3G4YJ38 A0A3G4YJ66 A0A0B5KTB0 A0A172MHM9 A0A3G3BTR3 A0A0B5KJX3 A0A3G4YJ20 A0A1L3KN56 A0A1L3KMX3 A0A1L3KN37 A0A1L3KN90 A0A3G5FMI8 A0A1L3KMV0 A0A3B1EJ88 A0A1L3KML0 A0A2S1ZPS1 C4NFK9 C4NFL5 A0A2Z4HF79 A0A2Z4HFC4 A0A2Z4HFJ7 A0A2Z4HF97 A0A2Z4HF69 A0A2Z4HFG4 A0A2Z4GIN2 A0A2Z4HFS7 A0A2H4YI41 A0A2Z4HFJ1 A0A2Z4HFC6 A0A0B5KEN7 A0A2Z4HFA6 A0A2Z4HFC7 A0A2Z4HFB8 A0A2Z4HFG9 A0A1L3KMU5 A0A0B5KRF3 A0A060DA11 A0A1L3KN23 A0A0X3PRB1 A0A067YEZ6 A0A1Z2RTE0 A0A0X3PYN5 A0A1Z2RSU8 A0A1Z2RTB4 A0A1Z2RT54 A0A1Z2RT41 A0A2Z4Z3N4 A0A0B5KTH4 A0A1B1FIU1 A0A0B5KK67
Ontologies
KEGG
GO
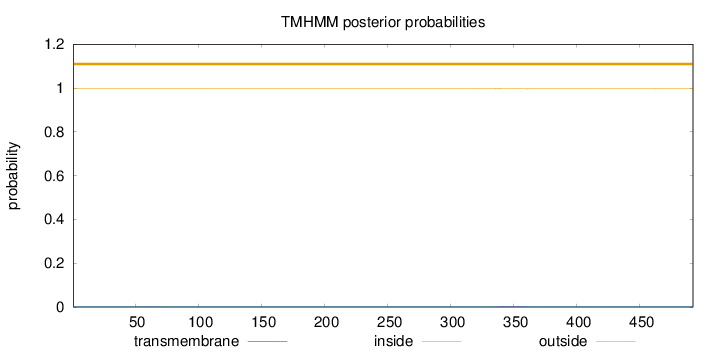
Topology
Subcellular location
Virion
Host cytoplasm
Host cytoplasm
Length:
492
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0661299999999999
Exp number, first 60 AAs:
0.00289
Total prob of N-in:
0.00105
outside
1 - 492
Population Genetic Test Statistics
Pi
230.627384
Theta
208.002971
Tajima's D
0.34314
CLR
249.860882
CSRT
0.463776811159442
Interpretation
Uncertain
