Gene
KWMTBOMO08476
Annotation
PREDICTED:_uncharacterized_protein_LOC105841278_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.723 Nuclear Reliability : 1.038
Sequence
CDS
ATGTTTAGGTGCATTCTGCTGGATCCAGAGCAACGTTCGCTGCAACTTATACTATTGCGCGACTCACCACATGAAAACATCAAGTGTATTGAACTAAATACAGTTACATATGGTTTTAAATGCTCAACCTATCTAGCCACTAGGTTTCTGACTGAACTAGCACACAGGTACGAGGCTGACTTCCCAGCGGCGTCTTTCATTCTGCAAAATCAGACATACTGCGACGACATTCTCACTTCCAGTAACCCTTTGGAGTCTTTATCTGAAATGAAAGATCAATTAATACAATTACTAGCTCTGGGAGGGTTTCAGGCACATAAATGGTCTTCAAATGCTCCTCAAATCCTGCAAAACATACCTAGAGATAAACAACATTTTGATGATGTCGATATACAAAAACAAAATTACTATATAAAAACATTAGGGGTCACATATAATACTAATACTGACACATTCAAAATATCAACTCCTAACCAACAGGGGCCCATGCCTCTCACAAAACGCGAAATCGTTAGTTTTGTCGCCAGATTTTACGACCCACTGGGTCTAGCTGGACCTATCACAGTTTCTGCAAAGATACTTATTCAGAAACTATGGGCAGCTCAAATCAACTGGGACTCACAATTGCCTAATGATCTTAAAACAGCTTGGTTAGAGTTCTATAATAATTTACATAGCATGCAGCCTATACACATCACTAGGAATGTCACTATGCAGCAGGCAGCCAGCTCACACCTGATCGGCTATGCAGATGCCAGCTGTGCAGCATATGGTTACTGTCTTTATCTGCGCGAAGTCGACAAGGTAGGTAAGGTCAAGGTCACTTTACTCTGTTCGAAATCAAGACTAGCTCCATTAAGCCAGAAACTAACAACACCTAGACTTGAGCTAAATGCCGTGCTTTTACTAGCTAAACTAGTACACAGAGTATATTCATTGTTATCTCTAAAAATACATATTGATGATGTATCTCTTTTCTCTGACTCTCAAATAGTCCTGGCGTGGTTTAAGCTAGATATTACAAAACTAAATGCCTATGTAGCAAACAGAGTAAAGACGATCTTGGAATTTACTAAAAATTTCCTCTGGACATACGTGAGGACGGGAGACAACCCTGCAGACTGTCTAAGCCGCGGCGCGCAGCCCAACGAGCTCGAGCACAACACGCTGTGGTGGCAGGGACCCGGTTACTTACATAACAGTAACTACTCTCCGACAAAAGTAAATATTAAAATATCAGATAAAATACCTGAGCTGAAGTCTGGTGATGCTGCTGATCCTCTGCCCCTCTCGAGTGCTATGGTCAAAAATTGA
Protein
MFRCILLDPEQRSLQLILLRDSPHENIKCIELNTVTYGFKCSTYLATRFLTELAHRYEADFPAASFILQNQTYCDDILTSSNPLESLSEMKDQLIQLLALGGFQAHKWSSNAPQILQNIPRDKQHFDDVDIQKQNYYIKTLGVTYNTNTDTFKISTPNQQGPMPLTKREIVSFVARFYDPLGLAGPITVSAKILIQKLWAAQINWDSQLPNDLKTAWLEFYNNLHSMQPIHITRNVTMQQAASSHLIGYADASCAAYGYCLYLREVDKVGKVKVTLLCSKSRLAPLSQKLTTPRLELNAVLLLAKLVHRVYSLLSLKIHIDDVSLFSDSQIVLAWFKLDITKLNAYVANRVKTILEFTKNFLWTYVRTGDNPADCLSRGAQPNELEHNTLWWQGPGYLHNSNYSPTKVNIKISDKIPELKSGDAADPLPLSSAMVKN
Summary
Similarity
Belongs to the synaptobrevin family.
Uniprot
A0A1Y1JX04
A0A3S2P870
A0A1Y1MY22
A0A0A9XBI3
A0A182H647
A0A224XGH3
+ More
A0A1B6MFR0 A0A2S2QWS1 A0A0J7KBV8 X1WVF9 A0A1Y1N4T1 A0A1Y1MF14 A0A0P6K153 A0A1Y1MIB7 A0A0P6J6G2 A0A1W7R6J0 A0A1B6KZV2 A0A1B6MUZ9 X1X9U0 A0A1W7R6J2 A0A224XG39 W8ANH7 A0A1Y1MTT1 A0A2L2YT61 A0A3S2TB02 A0A1W7R6N3 A0A2S2PNZ4 X1XET3 X1WKA2 A0A0J7N314 J9L9H5 A0A2S2NPW7 E9J9Y8 A0A1B6GGX5 A0A0J7K7S0 J9JUM9 W4VR47 A0A087UK63 A0A0J7JX13 J9LHB7 A0A2L2YSX3 X1XU46 X1X412 X1X8C2 A0A2A4IUY1 A0A0J7K907 A0A3L8DC69 J9KV52 A0A1W7R6A8 A0A0A9XDK6 A0A2A4IUL3 A0A0J7KGI8 A0A2S2NDE3 A0A2S2NZ32 X1WPD9 J9JUK4 K7JBZ1 A0A182I0G9 A0A023F096 A0A0J7K2F3 A0A1Y1LAD7 K7JBZ2 A0A0J7KBZ0 X1X4W4 A0A1W7R6I7 A0A2S2P1X6 A0A0J7MVH5 A0A1W7R6S6 A0A1Y1L369 A0A3Q0JNQ0 A0A0A9X2P1 W8BU98 A0A2S2QTX5 A0A0J7JWZ4 A0A1S3DCY4 X1WWR4 A0A0A9Z7S5 J9JUA7 A0A3Q0JHI8 A0A1B6GMH9 A0A1B6LDH0 J9L3C9 X1WNM7 A0A182YS31 X1WNT8 A0A1U8N896 A0A0J7K7L1 A0A2S2N8W8 A0A0A9W8L8 J9M7R2 J9K0E7 A0A146LBU6 A0A023EZC2 A0A2S2PC00 A0A023EZB2 A0A226D417 A0A437AV42 A0A3Q0JGS5 A0A0A9YD88 A0A0J7KB34
A0A1B6MFR0 A0A2S2QWS1 A0A0J7KBV8 X1WVF9 A0A1Y1N4T1 A0A1Y1MF14 A0A0P6K153 A0A1Y1MIB7 A0A0P6J6G2 A0A1W7R6J0 A0A1B6KZV2 A0A1B6MUZ9 X1X9U0 A0A1W7R6J2 A0A224XG39 W8ANH7 A0A1Y1MTT1 A0A2L2YT61 A0A3S2TB02 A0A1W7R6N3 A0A2S2PNZ4 X1XET3 X1WKA2 A0A0J7N314 J9L9H5 A0A2S2NPW7 E9J9Y8 A0A1B6GGX5 A0A0J7K7S0 J9JUM9 W4VR47 A0A087UK63 A0A0J7JX13 J9LHB7 A0A2L2YSX3 X1XU46 X1X412 X1X8C2 A0A2A4IUY1 A0A0J7K907 A0A3L8DC69 J9KV52 A0A1W7R6A8 A0A0A9XDK6 A0A2A4IUL3 A0A0J7KGI8 A0A2S2NDE3 A0A2S2NZ32 X1WPD9 J9JUK4 K7JBZ1 A0A182I0G9 A0A023F096 A0A0J7K2F3 A0A1Y1LAD7 K7JBZ2 A0A0J7KBZ0 X1X4W4 A0A1W7R6I7 A0A2S2P1X6 A0A0J7MVH5 A0A1W7R6S6 A0A1Y1L369 A0A3Q0JNQ0 A0A0A9X2P1 W8BU98 A0A2S2QTX5 A0A0J7JWZ4 A0A1S3DCY4 X1WWR4 A0A0A9Z7S5 J9JUA7 A0A3Q0JHI8 A0A1B6GMH9 A0A1B6LDH0 J9L3C9 X1WNM7 A0A182YS31 X1WNT8 A0A1U8N896 A0A0J7K7L1 A0A2S2N8W8 A0A0A9W8L8 J9M7R2 J9K0E7 A0A146LBU6 A0A023EZC2 A0A2S2PC00 A0A023EZB2 A0A226D417 A0A437AV42 A0A3Q0JGS5 A0A0A9YD88 A0A0J7KB34
Pubmed
EMBL
GEZM01103137
GEZM01103136
JAV51536.1
RSAL01000210
RVE44241.1
GEZM01018702
+ More
JAV90048.1 GBHO01029149 JAG14455.1 JXUM01113346 KQ565681 KXJ70776.1 GFTR01008866 JAW07560.1 GEBQ01005223 JAT34754.1 GGMS01012379 MBY81582.1 LBMM01009941 KMQ87772.1 ABLF02026240 ABLF02055027 GEZM01012999 JAV92824.1 GEZM01033155 JAV84392.1 GDUN01000087 JAN95832.1 GEZM01033158 GEZM01033157 JAV84390.1 GDUN01000088 JAN95831.1 GEHC01000877 JAV46768.1 GEBQ01022991 GEBQ01006185 JAT16986.1 JAT33792.1 GEBQ01007199 GEBQ01000222 JAT32778.1 JAT39755.1 ABLF02025690 GEHC01000894 JAV46751.1 GFTR01008874 JAW07552.1 GAMC01020552 JAB86003.1 GEZM01021505 JAV88969.1 IAAA01049856 LAA11259.1 RSAL01001808 RVE40681.1 GEHC01000865 JAV46780.1 GGMR01018506 MBY31125.1 ABLF02041762 ABLF02011640 ABLF02018832 ABLF02018833 ABLF02047353 ABLF02057892 ABLF02066963 LBMM01011051 KMQ87060.1 ABLF02008636 ABLF02065931 GGMR01006575 MBY19194.1 GL769615 EFZ10365.1 GECZ01008071 JAS61698.1 LBMM01012392 KMQ86241.1 ABLF02004035 ABLF02004036 ABLF02004037 ABLF02004038 ABLF02008018 ABLF02008020 ABLF02012635 ABLF02061171 GANO01004788 JAB55083.1 KK120200 KFM77752.1 LBMM01024134 KMQ82612.1 ABLF02022040 ABLF02056888 IAAA01040237 LAA10530.1 ABLF02003339 ABLF02058001 ABLF02008635 ABLF02023121 ABLF02065930 NWSH01006923 PCG63188.1 LBMM01011516 KMQ86784.1 QOIP01000010 RLU18065.1 ABLF02015610 ABLF02015611 ABLF02026843 ABLF02057498 ABLF02060119 ABLF02062416 ABLF02064495 GEHC01000946 JAV46699.1 GBHO01028449 JAG15155.1 PCG63186.1 LBMM01007859 KMQ89359.1 GGMR01002187 MBY14806.1 GGMR01009854 MBY22473.1 ABLF02006985 ABLF02039606 AAZX01023184 APCN01002077 GBBI01004276 JAC14436.1 LBMM01016286 KMQ84479.1 GEZM01066274 JAV68017.1 LBMM01009964 KMQ87756.1 ABLF02043036 GEHC01000869 JAV46776.1 GGMR01010723 MBY23342.1 LBMM01016285 KMQ84480.1 GEHC01000870 JAV46775.1 GEZM01066272 JAV68024.1 GBHO01028617 JAG14987.1 GAMC01009674 JAB96881.1 GGMS01011369 MBY80572.1 LBMM01024307 KMQ82592.1 ABLF02058750 GBHO01004196 JAG39408.1 ABLF02041650 GECZ01006169 JAS63600.1 GEBQ01018232 JAT21745.1 ABLF02042806 ABLF02009451 LBMM01012188 KMQ86362.1 GGMR01000961 MBY13580.1 GBHO01039838 GBHO01006034 JAG03766.1 JAG37570.1 ABLF02005251 ABLF02055296 GDHC01014057 JAQ04572.1 GBBI01004139 JAC14573.1 GGMR01014351 MBY26970.1 GBBI01004140 JAC14572.1 LNIX01000036 OXA39969.1 RSAL01000392 RVE41994.1 GBHO01020765 GBHO01014531 JAG22839.1 JAG29073.1 LBMM01010405 KMQ87487.1
JAV90048.1 GBHO01029149 JAG14455.1 JXUM01113346 KQ565681 KXJ70776.1 GFTR01008866 JAW07560.1 GEBQ01005223 JAT34754.1 GGMS01012379 MBY81582.1 LBMM01009941 KMQ87772.1 ABLF02026240 ABLF02055027 GEZM01012999 JAV92824.1 GEZM01033155 JAV84392.1 GDUN01000087 JAN95832.1 GEZM01033158 GEZM01033157 JAV84390.1 GDUN01000088 JAN95831.1 GEHC01000877 JAV46768.1 GEBQ01022991 GEBQ01006185 JAT16986.1 JAT33792.1 GEBQ01007199 GEBQ01000222 JAT32778.1 JAT39755.1 ABLF02025690 GEHC01000894 JAV46751.1 GFTR01008874 JAW07552.1 GAMC01020552 JAB86003.1 GEZM01021505 JAV88969.1 IAAA01049856 LAA11259.1 RSAL01001808 RVE40681.1 GEHC01000865 JAV46780.1 GGMR01018506 MBY31125.1 ABLF02041762 ABLF02011640 ABLF02018832 ABLF02018833 ABLF02047353 ABLF02057892 ABLF02066963 LBMM01011051 KMQ87060.1 ABLF02008636 ABLF02065931 GGMR01006575 MBY19194.1 GL769615 EFZ10365.1 GECZ01008071 JAS61698.1 LBMM01012392 KMQ86241.1 ABLF02004035 ABLF02004036 ABLF02004037 ABLF02004038 ABLF02008018 ABLF02008020 ABLF02012635 ABLF02061171 GANO01004788 JAB55083.1 KK120200 KFM77752.1 LBMM01024134 KMQ82612.1 ABLF02022040 ABLF02056888 IAAA01040237 LAA10530.1 ABLF02003339 ABLF02058001 ABLF02008635 ABLF02023121 ABLF02065930 NWSH01006923 PCG63188.1 LBMM01011516 KMQ86784.1 QOIP01000010 RLU18065.1 ABLF02015610 ABLF02015611 ABLF02026843 ABLF02057498 ABLF02060119 ABLF02062416 ABLF02064495 GEHC01000946 JAV46699.1 GBHO01028449 JAG15155.1 PCG63186.1 LBMM01007859 KMQ89359.1 GGMR01002187 MBY14806.1 GGMR01009854 MBY22473.1 ABLF02006985 ABLF02039606 AAZX01023184 APCN01002077 GBBI01004276 JAC14436.1 LBMM01016286 KMQ84479.1 GEZM01066274 JAV68017.1 LBMM01009964 KMQ87756.1 ABLF02043036 GEHC01000869 JAV46776.1 GGMR01010723 MBY23342.1 LBMM01016285 KMQ84480.1 GEHC01000870 JAV46775.1 GEZM01066272 JAV68024.1 GBHO01028617 JAG14987.1 GAMC01009674 JAB96881.1 GGMS01011369 MBY80572.1 LBMM01024307 KMQ82592.1 ABLF02058750 GBHO01004196 JAG39408.1 ABLF02041650 GECZ01006169 JAS63600.1 GEBQ01018232 JAT21745.1 ABLF02042806 ABLF02009451 LBMM01012188 KMQ86362.1 GGMR01000961 MBY13580.1 GBHO01039838 GBHO01006034 JAG03766.1 JAG37570.1 ABLF02005251 ABLF02055296 GDHC01014057 JAQ04572.1 GBBI01004139 JAC14573.1 GGMR01014351 MBY26970.1 GBBI01004140 JAC14572.1 LNIX01000036 OXA39969.1 RSAL01000392 RVE41994.1 GBHO01020765 GBHO01014531 JAG22839.1 JAG29073.1 LBMM01010405 KMQ87487.1
Proteomes
Pfam
Interpro
IPR008042
Retrotrans_Pao
+ More
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR021109 Peptidase_aspartic_dom_sf
IPR008737 Peptidase_asp_put
IPR040676 DUF5641
IPR005312 DUF1759
IPR000477 RT_dom
IPR001878 Znf_CCHC
IPR001995 Peptidase_A2_cat
IPR001969 Aspartic_peptidase_AS
IPR036291 NAD(P)-bd_dom_sf
IPR025398 DUF4371
IPR020831 GlycerAld/Erythrose_P_DH
IPR020828 GlycerAld_3-P_DH_NAD(P)-bd
IPR017907 Znf_RING_CS
IPR001606 ARID_dom
IPR006612 THAP_Znf
IPR036875 Znf_CCHC_sf
IPR015416 Znf_H2C2_histone_UAS-bd
IPR038563 Endonuclease_7_sf
IPR004211 Endonuclease_7
IPR011012 Longin-like_dom_sf
IPR010908 Longin_dom
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR021109 Peptidase_aspartic_dom_sf
IPR008737 Peptidase_asp_put
IPR040676 DUF5641
IPR005312 DUF1759
IPR000477 RT_dom
IPR001878 Znf_CCHC
IPR001995 Peptidase_A2_cat
IPR001969 Aspartic_peptidase_AS
IPR036291 NAD(P)-bd_dom_sf
IPR025398 DUF4371
IPR020831 GlycerAld/Erythrose_P_DH
IPR020828 GlycerAld_3-P_DH_NAD(P)-bd
IPR017907 Znf_RING_CS
IPR001606 ARID_dom
IPR006612 THAP_Znf
IPR036875 Znf_CCHC_sf
IPR015416 Znf_H2C2_histone_UAS-bd
IPR038563 Endonuclease_7_sf
IPR004211 Endonuclease_7
IPR011012 Longin-like_dom_sf
IPR010908 Longin_dom
SUPFAM
Gene 3D
ProteinModelPortal
A0A1Y1JX04
A0A3S2P870
A0A1Y1MY22
A0A0A9XBI3
A0A182H647
A0A224XGH3
+ More
A0A1B6MFR0 A0A2S2QWS1 A0A0J7KBV8 X1WVF9 A0A1Y1N4T1 A0A1Y1MF14 A0A0P6K153 A0A1Y1MIB7 A0A0P6J6G2 A0A1W7R6J0 A0A1B6KZV2 A0A1B6MUZ9 X1X9U0 A0A1W7R6J2 A0A224XG39 W8ANH7 A0A1Y1MTT1 A0A2L2YT61 A0A3S2TB02 A0A1W7R6N3 A0A2S2PNZ4 X1XET3 X1WKA2 A0A0J7N314 J9L9H5 A0A2S2NPW7 E9J9Y8 A0A1B6GGX5 A0A0J7K7S0 J9JUM9 W4VR47 A0A087UK63 A0A0J7JX13 J9LHB7 A0A2L2YSX3 X1XU46 X1X412 X1X8C2 A0A2A4IUY1 A0A0J7K907 A0A3L8DC69 J9KV52 A0A1W7R6A8 A0A0A9XDK6 A0A2A4IUL3 A0A0J7KGI8 A0A2S2NDE3 A0A2S2NZ32 X1WPD9 J9JUK4 K7JBZ1 A0A182I0G9 A0A023F096 A0A0J7K2F3 A0A1Y1LAD7 K7JBZ2 A0A0J7KBZ0 X1X4W4 A0A1W7R6I7 A0A2S2P1X6 A0A0J7MVH5 A0A1W7R6S6 A0A1Y1L369 A0A3Q0JNQ0 A0A0A9X2P1 W8BU98 A0A2S2QTX5 A0A0J7JWZ4 A0A1S3DCY4 X1WWR4 A0A0A9Z7S5 J9JUA7 A0A3Q0JHI8 A0A1B6GMH9 A0A1B6LDH0 J9L3C9 X1WNM7 A0A182YS31 X1WNT8 A0A1U8N896 A0A0J7K7L1 A0A2S2N8W8 A0A0A9W8L8 J9M7R2 J9K0E7 A0A146LBU6 A0A023EZC2 A0A2S2PC00 A0A023EZB2 A0A226D417 A0A437AV42 A0A3Q0JGS5 A0A0A9YD88 A0A0J7KB34
A0A1B6MFR0 A0A2S2QWS1 A0A0J7KBV8 X1WVF9 A0A1Y1N4T1 A0A1Y1MF14 A0A0P6K153 A0A1Y1MIB7 A0A0P6J6G2 A0A1W7R6J0 A0A1B6KZV2 A0A1B6MUZ9 X1X9U0 A0A1W7R6J2 A0A224XG39 W8ANH7 A0A1Y1MTT1 A0A2L2YT61 A0A3S2TB02 A0A1W7R6N3 A0A2S2PNZ4 X1XET3 X1WKA2 A0A0J7N314 J9L9H5 A0A2S2NPW7 E9J9Y8 A0A1B6GGX5 A0A0J7K7S0 J9JUM9 W4VR47 A0A087UK63 A0A0J7JX13 J9LHB7 A0A2L2YSX3 X1XU46 X1X412 X1X8C2 A0A2A4IUY1 A0A0J7K907 A0A3L8DC69 J9KV52 A0A1W7R6A8 A0A0A9XDK6 A0A2A4IUL3 A0A0J7KGI8 A0A2S2NDE3 A0A2S2NZ32 X1WPD9 J9JUK4 K7JBZ1 A0A182I0G9 A0A023F096 A0A0J7K2F3 A0A1Y1LAD7 K7JBZ2 A0A0J7KBZ0 X1X4W4 A0A1W7R6I7 A0A2S2P1X6 A0A0J7MVH5 A0A1W7R6S6 A0A1Y1L369 A0A3Q0JNQ0 A0A0A9X2P1 W8BU98 A0A2S2QTX5 A0A0J7JWZ4 A0A1S3DCY4 X1WWR4 A0A0A9Z7S5 J9JUA7 A0A3Q0JHI8 A0A1B6GMH9 A0A1B6LDH0 J9L3C9 X1WNM7 A0A182YS31 X1WNT8 A0A1U8N896 A0A0J7K7L1 A0A2S2N8W8 A0A0A9W8L8 J9M7R2 J9K0E7 A0A146LBU6 A0A023EZC2 A0A2S2PC00 A0A023EZB2 A0A226D417 A0A437AV42 A0A3Q0JGS5 A0A0A9YD88 A0A0J7KB34
Ontologies
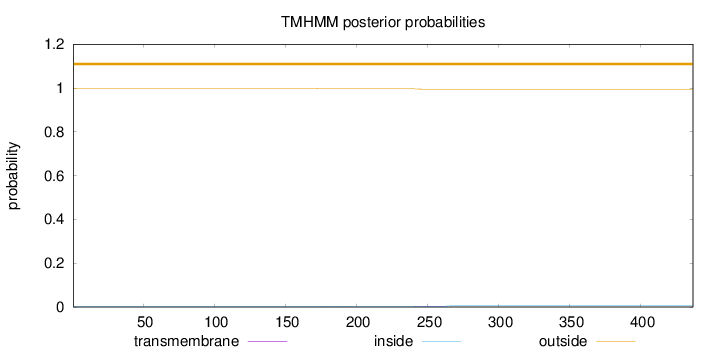
Topology
Length:
437
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0956900000000001
Exp number, first 60 AAs:
0.0008
Total prob of N-in:
0.00165
outside
1 - 437
Population Genetic Test Statistics
Pi
69.941864
Theta
176.289695
Tajima's D
-1.815481
CLR
1280.798329
CSRT
0.0260986950652467
Interpretation
Uncertain
