Gene
KWMTBOMO08468
Pre Gene Modal
BGIBMGA009425
Annotation
PREDICTED:_brain_tumor_protein_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.446 Nuclear Reliability : 1.502
Sequence
CDS
ATGGCCTCACGCACCCCGTCCCTTGAGTCGCTGCCCGGTGCCAACTCAATAGGTTCGCTGGAGCGTGGCTCCATATCGCCCCTCACTCTCAGCGGTTCATCACCCCCTGCGAGCGACTCCGCCGTATGTGATTTACGCGACTTCGACGGACTCGATACCACTTGCGCCATCTGCCGTGAGACATTCATCGACCCCAAAGTACTTAACTGCTTTCATACTTTTTGCCGGGCTTGCCTGGACAGAGAACAGACCCACCCGGACAGAGTAACGTGCGTTACTTGTCGTGTTGACACTCAATTGCCACCCGGTGGTGTTGACAGCCTTCTTACGAATTTAGTGATCGCCGCTGCTGTCGACCAAGACGCTGAGCTGCTCTCATCTGGGCGACAGACAAGCTCTCCCTTGGCTCGTTGCACCGGCTGCAAGTCCAAGGAATCTGACGCCGTAGCGCGTTGTGTTGATTGCGCAAACTACCTCTGCCCCAACTGCGTCATGGCACACCAATTCATGCATTGCTTCGAAGGCCATCGAGTTCTTGCATTCACCGACATGAAAGACGACAAAGGCATACTGGGGACTACTATTACGACAAGCGGAGAGAAAACCGCATTCTGCCCAAGACACAAAAATGACATCTTGAAGTATTACTGCCGTACGTGCTCCGTGCCGGTGTGCAAGGAATGCACCATTATAGAGCATCCAGCGGCTCTGCACGACTGCGAGCATCTCTCTGACGCCGGTCCCAAACAGTTGGAGCTCATGCAGCAAGCGGTAAACGAAGCTAAGACCCGCGCAACTGAAATAAGGCACGTGGTCAAGACAGTAGAGCACGCCGCTGGAAAATTACAAGTTCAGTATCACAAGGCACAGAATGAGATAAACGACACTTTCCAGTTCTATCGCTCCATGTTGGAAGAACGAAAACAAGAACTGCTAAAAGAGCTAGAAAGTGTATTTTCTACCAAACAAATTGCCCTGACAGTGGTTGGACAAAAGGCGCAGGAAACTGTAGATAAGATTTATCAGACATGCGACTTTGTCGAGAAACTGACGAAATGCGCCAACATCGCTGAGATCCTTATGTTCAGGAAACTTCTCGATACCAAACTGCAATCCTTGATGAGCTCTAACCCCGAACAAAGCGTACAGACAGCATGCGAACTGGAATTTGTTTCCAACTACCAAGCCATCCAGGTAGGTGTACGAAACACTTTCGGGTACGTACGCTCAAGCTCAGAAGCAAACGTCGGTCCGACTAAACAACCACCGATCGCTCGTCCAACAAACGGATCCCTGTTGAATGGAGGCTCATCATCCAGCAGCAGCAGCGTGAACGGAAGCTCTGGAAGCTTGAACGGTGGCATTCATCTACCTACCGGTTTGAACGGAGTTCTGGATCGTCCATTTACTAATGGGCTTCTAGGGCCTACTAGCCAGTCTACTTCGCCATTCGAATCTAATTTGATTTCAAAACGATTCAACAGCGTCAACAGCCTCGGGCCTTTCTCTGCTGCTATCGGAGATATGAATCTAAACGGAATTAATCCATACGAAAAATGGTCCAACGGTGGTTGTGATGCCCTGTTTCCACCAACCACAACTGACCCATACTCACTGACTGCTGTGGCACATACCGATCCTCTGCTAGACCTAACAAACAAATTGATTTCCACTGCTATGTTCCCGCCTAAATCTCAAATCAAGCGCCAAAAAATGATTTACCATTGCAAATTCGGAGAGTTTGGCGTTATGGAAGGCCAATTCACGGAACCCAGCGGTGTCGCGGTCAATGCTCAGAACGATATCATTGTTGCCGATACAAACAACCACCGCATTCAGATCTTCGACAAAGAAGGTCGTTTCAAATTCCAATTCGGGGAATGCGGCAAGCGTGACGGCCAGTTACTGTATCCTAACCGCGTCGCTGTTGTTCGCACATCAGGAGACATAATTGTGACAGAACGATCGCCAACACACCAGATCCAGATCTATAACCAATACGGGCAGTTTGTACGCAAATTCGGGGCAAACATCTTGCAACATCCCCGCGGAGTCACCGTCGACAACAAGGGCCGTATCGTTGTCGTCGAATGCAAAGTAATGCGGGTTATCATTTTCGATCAAGTGGGCAACGTACTACAGAAGTTTGGCTGCTCTAAGCACTTGGAATTCCCCAACGGCGTCGTCGTTAATGATAAGCAGGAGATTTTCATCAGTGACAATCGCGCACATTGCGTCAAAGTATTCAATTATGACGGCATTTACTTGCGTCAGATCGGGGGCGAAGGTGTCACCAATTATCCGATCGGCGTTGGCATCAACGCAGCTGGTGAGATCTTGATCGCCGATAACCACAACAACTTCAATCTGACGATCTTCACGCAGGAGGGCCAACTCGTATCTGCCCTCGAAAGTAAGGTCAAACACGCTCAATGCTTCGACGTAGCGCTCATGGACGACGGCTCAGTTGTACTTGCGAGCAAGGACTACCGGCTGTACATCTACCGCTACGTACAAGTGCCCCCAATAGGCATGTGA
Protein
MASRTPSLESLPGANSIGSLERGSISPLTLSGSSPPASDSAVCDLRDFDGLDTTCAICRETFIDPKVLNCFHTFCRACLDREQTHPDRVTCVTCRVDTQLPPGGVDSLLTNLVIAAAVDQDAELLSSGRQTSSPLARCTGCKSKESDAVARCVDCANYLCPNCVMAHQFMHCFEGHRVLAFTDMKDDKGILGTTITTSGEKTAFCPRHKNDILKYYCRTCSVPVCKECTIIEHPAALHDCEHLSDAGPKQLELMQQAVNEAKTRATEIRHVVKTVEHAAGKLQVQYHKAQNEINDTFQFYRSMLEERKQELLKELESVFSTKQIALTVVGQKAQETVDKIYQTCDFVEKLTKCANIAEILMFRKLLDTKLQSLMSSNPEQSVQTACELEFVSNYQAIQVGVRNTFGYVRSSSEANVGPTKQPPIARPTNGSLLNGGSSSSSSSVNGSSGSLNGGIHLPTGLNGVLDRPFTNGLLGPTSQSTSPFESNLISKRFNSVNSLGPFSAAIGDMNLNGINPYEKWSNGGCDALFPPTTTDPYSLTAVAHTDPLLDLTNKLISTAMFPPKSQIKRQKMIYHCKFGEFGVMEGQFTEPSGVAVNAQNDIIVADTNNHRIQIFDKEGRFKFQFGECGKRDGQLLYPNRVAVVRTSGDIIVTERSPTHQIQIYNQYGQFVRKFGANILQHPRGVTVDNKGRIVVVECKVMRVIIFDQVGNVLQKFGCSKHLEFPNGVVVNDKQEIFISDNRAHCVKVFNYDGIYLRQIGGEGVTNYPIGVGINAAGEILIADNHNNFNLTIFTQEGQLVSALESKVKHAQCFDVALMDDGSVVLASKDYRLYIYRYVQVPPIGM
Summary
Uniprot
H9JIS4
A0A2A4JPN7
A0A2H1VUN1
A0A212EU52
A0A3S2P147
A0A0N1PJZ9
+ More
A0A194Q7Q1 A0A0L7LN57 A0A1E1WUQ7 D2A0S0 A0A1W4WQF3 A0A1B6CC71 A0A1Y1LDM3 A0A2J7PZH7 N6TP29 U4UF78 A0A146M6Y2 A0A0A9WQ39 A0A0K8SLN2 A0A2R7W009 A0A1B6FGV4 A0A1B6LT16 A0A1B6K8W8 A0A0C9RSX1 T1HT84 A0A0L7QW79 A0A023F3H9 A0A2S2Q7T9 E0VBI3 A0A154PQ78 J9K3A6 A0A2H8TMT5 A0A2S2PU95 A0A310SS56 A0A0M8ZRW1 A0A2A3E193 A0A087ZVR7 K7ISI8 A0A067RBK6 A0A026WPE3 A0A3L8DXV6 A0A158NVB9 A0A3Q0JLM8 A0A0J7KYR9 A0A195F2S3 T1IPK5 A0A195C8Q6 A0A087T8B3 A0A2P6KUR7 A0A444SEJ3 A0A1S3JML1 A0A151XJG5 A0A1B0CY34 A0A1E1XEA0 L7MIF0 R7VKA8 A0A2R5LGZ1 A0A293M5D9 K1PZL0 A0A2P6KT45 A0A210PHL4 A0A336MNA1 A0A1Q3EVJ9 A0A195BFK9 Q17BL6 A0A2H5BFB6 B0XCV6 A0A195EKS6 A0A0L8HCK1 A0A182H4D2 E9I8W7 V4CDL6 E2ATR8 E2C9S7 A0A3S3PEW7 A0A443SQ59 Q7PMI8 A0A162SFW8 A0A0P6BLM7 A0A0P5VB53 A0A1J1ILX8 A0A0P5LTT3 A0A087T3M3 A0A0P6H0B1 A0A0P5F121 E9FRJ8 A0A077Z4J3 A0A1A9XL02 A0A1I8MJ76 L0PRP0 A0A1I8PSJ1 W8AGV2 A0A1B0FEZ7 A0A1B0A544 A0A0L0BQN3 A0A1A9W051 A0A0A1WSI9
A0A194Q7Q1 A0A0L7LN57 A0A1E1WUQ7 D2A0S0 A0A1W4WQF3 A0A1B6CC71 A0A1Y1LDM3 A0A2J7PZH7 N6TP29 U4UF78 A0A146M6Y2 A0A0A9WQ39 A0A0K8SLN2 A0A2R7W009 A0A1B6FGV4 A0A1B6LT16 A0A1B6K8W8 A0A0C9RSX1 T1HT84 A0A0L7QW79 A0A023F3H9 A0A2S2Q7T9 E0VBI3 A0A154PQ78 J9K3A6 A0A2H8TMT5 A0A2S2PU95 A0A310SS56 A0A0M8ZRW1 A0A2A3E193 A0A087ZVR7 K7ISI8 A0A067RBK6 A0A026WPE3 A0A3L8DXV6 A0A158NVB9 A0A3Q0JLM8 A0A0J7KYR9 A0A195F2S3 T1IPK5 A0A195C8Q6 A0A087T8B3 A0A2P6KUR7 A0A444SEJ3 A0A1S3JML1 A0A151XJG5 A0A1B0CY34 A0A1E1XEA0 L7MIF0 R7VKA8 A0A2R5LGZ1 A0A293M5D9 K1PZL0 A0A2P6KT45 A0A210PHL4 A0A336MNA1 A0A1Q3EVJ9 A0A195BFK9 Q17BL6 A0A2H5BFB6 B0XCV6 A0A195EKS6 A0A0L8HCK1 A0A182H4D2 E9I8W7 V4CDL6 E2ATR8 E2C9S7 A0A3S3PEW7 A0A443SQ59 Q7PMI8 A0A162SFW8 A0A0P6BLM7 A0A0P5VB53 A0A1J1ILX8 A0A0P5LTT3 A0A087T3M3 A0A0P6H0B1 A0A0P5F121 E9FRJ8 A0A077Z4J3 A0A1A9XL02 A0A1I8MJ76 L0PRP0 A0A1I8PSJ1 W8AGV2 A0A1B0FEZ7 A0A1B0A544 A0A0L0BQN3 A0A1A9W051 A0A0A1WSI9
Pubmed
19121390
22118469
26354079
26227816
18362917
19820115
+ More
28004739 23537049 26823975 25401762 25474469 20566863 20075255 24845553 24508170 30249741 21347285 28503490 25576852 23254933 22992520 28812685 17510324 29228898 26483478 21282665 20798317 12364791 21292972 25315136 24495485 26108605 25830018
28004739 23537049 26823975 25401762 25474469 20566863 20075255 24845553 24508170 30249741 21347285 28503490 25576852 23254933 22992520 28812685 17510324 29228898 26483478 21282665 20798317 12364791 21292972 25315136 24495485 26108605 25830018
EMBL
BABH01033002
NWSH01000871
PCG73779.1
ODYU01004547
SOQ44551.1
AGBW02012456
+ More
OWR45023.1 RSAL01000059 RVE49699.1 KQ460144 KPJ17459.1 KQ459337 KPJ01419.1 JTDY01000504 KOB76865.1 GDQN01000336 JAT90718.1 KQ971338 EFA02902.1 GEDC01030393 GEDC01026240 GEDC01021949 GEDC01020577 GEDC01014851 GEDC01007992 GEDC01006621 GEDC01002285 JAS06905.1 JAS11058.1 JAS15349.1 JAS16721.1 JAS22447.1 JAS29306.1 JAS30677.1 JAS35013.1 GEZM01058349 GEZM01058348 JAV71759.1 NEVH01020335 PNF21741.1 APGK01014745 APGK01020092 KB740160 KB739406 ENN81214.1 ENN82284.1 KB632077 ERL88570.1 GDHC01003837 JAQ14792.1 GBHO01034068 GDHC01013398 JAG09536.1 JAQ05231.1 GBRD01011656 JAG54168.1 KK854193 PTY12909.1 GECZ01020333 JAS49436.1 GEBQ01013146 JAT26831.1 GEBQ01032081 JAT07896.1 GBYB01010595 GBYB01010596 JAG80362.1 JAG80363.1 ACPB03002583 KQ414716 KOC62878.1 GBBI01002804 JAC15908.1 GGMS01004602 MBY73805.1 DS235030 EEB10739.1 KQ435005 KZC13408.1 ABLF02032120 GFXV01003485 MBW15290.1 GGMR01020421 MBY33040.1 KQ760662 OAD59519.1 KQ435927 KOX68383.1 KZ288461 PBC25475.1 KK852566 KDR21251.1 KK107144 EZA57526.1 QOIP01000003 RLU25281.1 ADTU01027066 LBMM01001987 KMQ95443.1 KQ981856 KYN34477.1 JH431263 KQ978081 KYM97085.1 KK113914 KFM61352.1 MWRG01004978 PRD30076.1 SAUD01039947 RXG51829.1 KQ982074 KYQ60529.1 AJWK01035350 GFAC01001772 JAT97416.1 GACK01001457 JAA63577.1 AMQN01017368 KB293041 ELU16635.1 GGLE01004660 MBY08786.1 GFWV01010797 MAA35526.1 JH816071 EKC24514.1 MWRG01005659 PRD29510.1 NEDP02076690 OWF35984.1 UFQT01001452 SSX30619.1 GFDL01015711 JAV19334.1 KQ976490 KYM83383.1 CH477320 EAT43650.1 MG197671 AUG84421.1 DS232721 EDS45123.1 KQ978782 KYN28474.1 KQ418549 KOF86907.1 JXUM01004067 JXUM01004068 JXUM01004069 JXUM01004070 JXUM01005781 KQ560201 KXJ83877.1 GL761662 EFZ22979.1 KB200869 ESP00015.1 GL442691 EFN63151.1 GL453897 EFN75317.1 NCKU01000009 RWS17996.1 NCKV01000838 RWS29605.1 AAAB01008980 EAA13775.5 LRGB01000027 KZS21263.1 GDIP01012921 JAM90794.1 GDIP01102085 JAM01630.1 CVRI01000054 CRL00754.1 GDIQ01165370 JAK86355.1 KK113255 KFM59712.1 GDIQ01036666 JAN58071.1 GDIQ01265074 JAJ86650.1 GL732523 EFX90185.1 HG805952 CDW55397.1 HE971733 CCK33022.1 GAMC01018941 GAMC01018940 JAB87615.1 CCAG010002751 JRES01001517 KNC22303.1 GBXI01012914 JAD01378.1
OWR45023.1 RSAL01000059 RVE49699.1 KQ460144 KPJ17459.1 KQ459337 KPJ01419.1 JTDY01000504 KOB76865.1 GDQN01000336 JAT90718.1 KQ971338 EFA02902.1 GEDC01030393 GEDC01026240 GEDC01021949 GEDC01020577 GEDC01014851 GEDC01007992 GEDC01006621 GEDC01002285 JAS06905.1 JAS11058.1 JAS15349.1 JAS16721.1 JAS22447.1 JAS29306.1 JAS30677.1 JAS35013.1 GEZM01058349 GEZM01058348 JAV71759.1 NEVH01020335 PNF21741.1 APGK01014745 APGK01020092 KB740160 KB739406 ENN81214.1 ENN82284.1 KB632077 ERL88570.1 GDHC01003837 JAQ14792.1 GBHO01034068 GDHC01013398 JAG09536.1 JAQ05231.1 GBRD01011656 JAG54168.1 KK854193 PTY12909.1 GECZ01020333 JAS49436.1 GEBQ01013146 JAT26831.1 GEBQ01032081 JAT07896.1 GBYB01010595 GBYB01010596 JAG80362.1 JAG80363.1 ACPB03002583 KQ414716 KOC62878.1 GBBI01002804 JAC15908.1 GGMS01004602 MBY73805.1 DS235030 EEB10739.1 KQ435005 KZC13408.1 ABLF02032120 GFXV01003485 MBW15290.1 GGMR01020421 MBY33040.1 KQ760662 OAD59519.1 KQ435927 KOX68383.1 KZ288461 PBC25475.1 KK852566 KDR21251.1 KK107144 EZA57526.1 QOIP01000003 RLU25281.1 ADTU01027066 LBMM01001987 KMQ95443.1 KQ981856 KYN34477.1 JH431263 KQ978081 KYM97085.1 KK113914 KFM61352.1 MWRG01004978 PRD30076.1 SAUD01039947 RXG51829.1 KQ982074 KYQ60529.1 AJWK01035350 GFAC01001772 JAT97416.1 GACK01001457 JAA63577.1 AMQN01017368 KB293041 ELU16635.1 GGLE01004660 MBY08786.1 GFWV01010797 MAA35526.1 JH816071 EKC24514.1 MWRG01005659 PRD29510.1 NEDP02076690 OWF35984.1 UFQT01001452 SSX30619.1 GFDL01015711 JAV19334.1 KQ976490 KYM83383.1 CH477320 EAT43650.1 MG197671 AUG84421.1 DS232721 EDS45123.1 KQ978782 KYN28474.1 KQ418549 KOF86907.1 JXUM01004067 JXUM01004068 JXUM01004069 JXUM01004070 JXUM01005781 KQ560201 KXJ83877.1 GL761662 EFZ22979.1 KB200869 ESP00015.1 GL442691 EFN63151.1 GL453897 EFN75317.1 NCKU01000009 RWS17996.1 NCKV01000838 RWS29605.1 AAAB01008980 EAA13775.5 LRGB01000027 KZS21263.1 GDIP01012921 JAM90794.1 GDIP01102085 JAM01630.1 CVRI01000054 CRL00754.1 GDIQ01165370 JAK86355.1 KK113255 KFM59712.1 GDIQ01036666 JAN58071.1 GDIQ01265074 JAJ86650.1 GL732523 EFX90185.1 HG805952 CDW55397.1 HE971733 CCK33022.1 GAMC01018941 GAMC01018940 JAB87615.1 CCAG010002751 JRES01001517 KNC22303.1 GBXI01012914 JAD01378.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000283053
UP000053240
UP000053268
+ More
UP000037510 UP000007266 UP000192223 UP000235965 UP000019118 UP000030742 UP000015103 UP000053825 UP000009046 UP000076502 UP000007819 UP000053105 UP000242457 UP000005203 UP000002358 UP000027135 UP000053097 UP000279307 UP000005205 UP000079169 UP000036403 UP000078541 UP000078542 UP000054359 UP000288706 UP000085678 UP000075809 UP000092461 UP000014760 UP000005408 UP000242188 UP000078540 UP000008820 UP000002320 UP000078492 UP000053454 UP000069940 UP000249989 UP000030746 UP000000311 UP000008237 UP000285301 UP000288716 UP000007062 UP000076858 UP000183832 UP000000305 UP000030665 UP000092443 UP000095301 UP000095300 UP000092444 UP000092445 UP000037069 UP000091820
UP000037510 UP000007266 UP000192223 UP000235965 UP000019118 UP000030742 UP000015103 UP000053825 UP000009046 UP000076502 UP000007819 UP000053105 UP000242457 UP000005203 UP000002358 UP000027135 UP000053097 UP000279307 UP000005205 UP000079169 UP000036403 UP000078541 UP000078542 UP000054359 UP000288706 UP000085678 UP000075809 UP000092461 UP000014760 UP000005408 UP000242188 UP000078540 UP000008820 UP000002320 UP000078492 UP000053454 UP000069940 UP000249989 UP000030746 UP000000311 UP000008237 UP000285301 UP000288716 UP000007062 UP000076858 UP000183832 UP000000305 UP000030665 UP000092443 UP000095301 UP000095300 UP000092444 UP000092445 UP000037069 UP000091820
Pfam
Interpro
IPR013083
Znf_RING/FYVE/PHD
+ More
IPR011042 6-blade_b-propeller_TolB-like
IPR017907 Znf_RING_CS
IPR001258 NHL_repeat
IPR001841 Znf_RING
IPR000315 Znf_B-box
IPR027370 Znf-RING_LisH
IPR003649 Bbox_C
IPR013017 NHL_repeat_subgr
IPR013087 Znf_C2H2_type
IPR002048 EF_hand_dom
IPR018247 EF_Hand_1_Ca_BS
IPR018957 Znf_C3HC4_RING-type
IPR040100 RNF208_RING-HC
IPR029058 AB_hydrolase
IPR019149 ABHD18
IPR003656 Znf_BED
IPR037220 BED_dom
IPR036236 Znf_C2H2_sf
IPR032041 Cdc73_N
IPR031336 CDC73_C
IPR038103 CDC73_C_sf
IPR011042 6-blade_b-propeller_TolB-like
IPR017907 Znf_RING_CS
IPR001258 NHL_repeat
IPR001841 Znf_RING
IPR000315 Znf_B-box
IPR027370 Znf-RING_LisH
IPR003649 Bbox_C
IPR013017 NHL_repeat_subgr
IPR013087 Znf_C2H2_type
IPR002048 EF_hand_dom
IPR018247 EF_Hand_1_Ca_BS
IPR018957 Znf_C3HC4_RING-type
IPR040100 RNF208_RING-HC
IPR029058 AB_hydrolase
IPR019149 ABHD18
IPR003656 Znf_BED
IPR037220 BED_dom
IPR036236 Znf_C2H2_sf
IPR032041 Cdc73_N
IPR031336 CDC73_C
IPR038103 CDC73_C_sf
Gene 3D
ProteinModelPortal
H9JIS4
A0A2A4JPN7
A0A2H1VUN1
A0A212EU52
A0A3S2P147
A0A0N1PJZ9
+ More
A0A194Q7Q1 A0A0L7LN57 A0A1E1WUQ7 D2A0S0 A0A1W4WQF3 A0A1B6CC71 A0A1Y1LDM3 A0A2J7PZH7 N6TP29 U4UF78 A0A146M6Y2 A0A0A9WQ39 A0A0K8SLN2 A0A2R7W009 A0A1B6FGV4 A0A1B6LT16 A0A1B6K8W8 A0A0C9RSX1 T1HT84 A0A0L7QW79 A0A023F3H9 A0A2S2Q7T9 E0VBI3 A0A154PQ78 J9K3A6 A0A2H8TMT5 A0A2S2PU95 A0A310SS56 A0A0M8ZRW1 A0A2A3E193 A0A087ZVR7 K7ISI8 A0A067RBK6 A0A026WPE3 A0A3L8DXV6 A0A158NVB9 A0A3Q0JLM8 A0A0J7KYR9 A0A195F2S3 T1IPK5 A0A195C8Q6 A0A087T8B3 A0A2P6KUR7 A0A444SEJ3 A0A1S3JML1 A0A151XJG5 A0A1B0CY34 A0A1E1XEA0 L7MIF0 R7VKA8 A0A2R5LGZ1 A0A293M5D9 K1PZL0 A0A2P6KT45 A0A210PHL4 A0A336MNA1 A0A1Q3EVJ9 A0A195BFK9 Q17BL6 A0A2H5BFB6 B0XCV6 A0A195EKS6 A0A0L8HCK1 A0A182H4D2 E9I8W7 V4CDL6 E2ATR8 E2C9S7 A0A3S3PEW7 A0A443SQ59 Q7PMI8 A0A162SFW8 A0A0P6BLM7 A0A0P5VB53 A0A1J1ILX8 A0A0P5LTT3 A0A087T3M3 A0A0P6H0B1 A0A0P5F121 E9FRJ8 A0A077Z4J3 A0A1A9XL02 A0A1I8MJ76 L0PRP0 A0A1I8PSJ1 W8AGV2 A0A1B0FEZ7 A0A1B0A544 A0A0L0BQN3 A0A1A9W051 A0A0A1WSI9
A0A194Q7Q1 A0A0L7LN57 A0A1E1WUQ7 D2A0S0 A0A1W4WQF3 A0A1B6CC71 A0A1Y1LDM3 A0A2J7PZH7 N6TP29 U4UF78 A0A146M6Y2 A0A0A9WQ39 A0A0K8SLN2 A0A2R7W009 A0A1B6FGV4 A0A1B6LT16 A0A1B6K8W8 A0A0C9RSX1 T1HT84 A0A0L7QW79 A0A023F3H9 A0A2S2Q7T9 E0VBI3 A0A154PQ78 J9K3A6 A0A2H8TMT5 A0A2S2PU95 A0A310SS56 A0A0M8ZRW1 A0A2A3E193 A0A087ZVR7 K7ISI8 A0A067RBK6 A0A026WPE3 A0A3L8DXV6 A0A158NVB9 A0A3Q0JLM8 A0A0J7KYR9 A0A195F2S3 T1IPK5 A0A195C8Q6 A0A087T8B3 A0A2P6KUR7 A0A444SEJ3 A0A1S3JML1 A0A151XJG5 A0A1B0CY34 A0A1E1XEA0 L7MIF0 R7VKA8 A0A2R5LGZ1 A0A293M5D9 K1PZL0 A0A2P6KT45 A0A210PHL4 A0A336MNA1 A0A1Q3EVJ9 A0A195BFK9 Q17BL6 A0A2H5BFB6 B0XCV6 A0A195EKS6 A0A0L8HCK1 A0A182H4D2 E9I8W7 V4CDL6 E2ATR8 E2C9S7 A0A3S3PEW7 A0A443SQ59 Q7PMI8 A0A162SFW8 A0A0P6BLM7 A0A0P5VB53 A0A1J1ILX8 A0A0P5LTT3 A0A087T3M3 A0A0P6H0B1 A0A0P5F121 E9FRJ8 A0A077Z4J3 A0A1A9XL02 A0A1I8MJ76 L0PRP0 A0A1I8PSJ1 W8AGV2 A0A1B0FEZ7 A0A1B0A544 A0A0L0BQN3 A0A1A9W051 A0A0A1WSI9
PDB
4ZLR
E-value=1.9969e-162,
Score=1472
Ontologies
GO
Topology
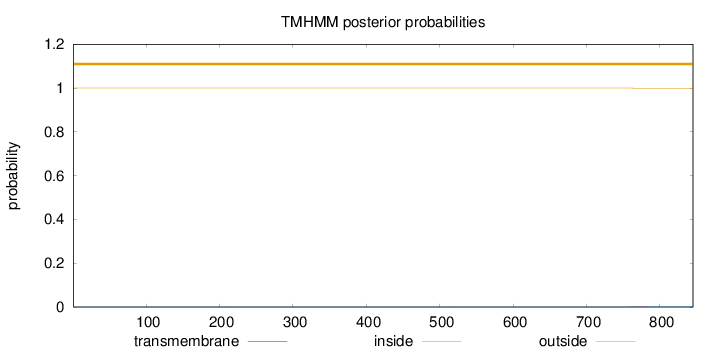
Length:
845
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01605
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00005
outside
1 - 845
Population Genetic Test Statistics
Pi
231.15826
Theta
200.589501
Tajima's D
0.479693
CLR
6.536535
CSRT
0.508224588770561
Interpretation
Uncertain
