Gene
KWMTBOMO08467
Pre Gene Modal
BGIBMGA009392
Annotation
Uncharacterized_protein_OBRU01_04785_[Operophtera_brumata]
Location in the cell
Cytoplasmic Reliability : 3.692
Sequence
CDS
ATGGGGGGCTTAACATTCGAAGGCGCATTGAAAAATATCAGTCACATCCAGCTTGAATTTATCAAGGAGGTCCTCGAGAAAAGGGGGTACAATGACAGAGTGGTTCATATAGAGGCGGTCGGGGCGGCCGGAGACAACTTCATCGCCAACGTCAAAAGGATAACAGTCGATGGCGAGAATGGCCCCTTCAAAATGATCGCTAAAATCGCCCCTCAAAATGAAAACGTTAGGATGGCCACACAAACACAGGTCTTATTTGGCAACGAACACCACATGTACACTGTTGTTTTGCCGAAATATCAGCAGCTAGAAGAAGAAGCAGAGATTCCTAATGACGACAGATTCAGATTTGCCGAATGCTATGGCTCAAGCACTCAAGTCCCGCATGAAGTGATATTACTAGAAGATCTACTGGTGCCGGGTTTTAAGATGCTGGACAGGTTCACGTCTCTATCTGACGAATGCATTCGGTCCACGCTCAAAAACTTCGCAAAGTTCCACGCTCTCTCGTTCACGCTTAAAAATAAACAGCCAGAAACCTTTAATTCGCTGAAGAGTAAATTATTCGATATGTGGAGTCACATGGATAGCTCAGCTGATATATACTTCGGAAAACTAGAAGCTGGTGCTGAGCAGCTCGTGGAGGGCGAAGAAAGGAAGAATATAATCAGGGGTGCCATTGGAAAGGCGCTTTCTATGGCACGAAAAATTGCTCAAGTCGAATCAGGGTCAAAATATTTCGTCATTCAACAAGGAGACTCGTGGACTAACAACATAATGTTCAGATATGATTGA
Protein
MGGLTFEGALKNISHIQLEFIKEVLEKRGYNDRVVHIEAVGAAGDNFIANVKRITVDGENGPFKMIAKIAPQNENVRMATQTQVLFGNEHHMYTVVLPKYQQLEEEAEIPNDDRFRFAECYGSSTQVPHEVILLEDLLVPGFKMLDRFTSLSDECIRSTLKNFAKFHALSFTLKNKQPETFNSLKSKLFDMWSHMDSSADIYFGKLEAGAEQLVEGEERKNIIRGAIGKALSMARKIAQVESGSKYFVIQQGDSWTNNIMFRYD
Summary
Uniprot
H9JIP2
A0A0L7LNI5
A0A0L7LN80
A0A2A4JDG0
A0A2H1X0D0
A0A2H1X0F4
+ More
A0A2A4JCA9 A0A2H1V3S2 A0A2A4K0J1 A0A2A4JV80 A0A2H1WA95 A0A2A4K1F1 A0A2W1BTI3 A0A2A4JBZ6 A0A2A4JX92 A0A2H1X0L3 A0A2W1BJJ0 A0A1E1WL08 A0A2A4JDV8 A0A3S2LB02 A0A2A4JDX0 A0A2A4JCM6 A0A2A4JCW2 A0A1E1WU33 A0A2A4JD29 A0A2H1WAI4 A0A2A4JCB8 A0A2A4JDU6 A0A0L7LNR9 A0A0N1IPP3 A0A194QD90 A0A0N1PHM5 A0A194Q7Q6 A0A194Q7A5 A0A0N1I9B8 A0A1E1WV54 A0A1E1WR59 A0A385GMR9 A0A1J1HIE9 U4TRU6 N6UJ02 U4UEL5 B3MWM8 B4NER2 A0A1W4UE05 Q9VYW8 A0A0A1XLB8 B4M238 A0A1Y1NAC9 U5ET31 A0A067QLU2 B4PXW6 B3NUB4 A0A1Y1LJQ0 A0A1J1HKX2 A0A2Z6FLE8 B4H7D8 A0A0K8TRH4 A0A1Q3G4W6 A0A3S2TJN1 Q29G00 Q7KV34 A0A1Q3G4V1 A0A212EH39 D6WTH0 A0A139WE97 A0A212EL62 A0A2P8YX69 B4IK36 A0A182VYD7 A0A212EPE1 A0A1W4WLY4 A0A0N1PEQ1 A0A2S2PG70 F6J1N0 A0A212FGC4 F6JLX3 H9JWK5 A9YGE4 A9YGD5 B0XJU1 F6JGA3
A0A2A4JCA9 A0A2H1V3S2 A0A2A4K0J1 A0A2A4JV80 A0A2H1WA95 A0A2A4K1F1 A0A2W1BTI3 A0A2A4JBZ6 A0A2A4JX92 A0A2H1X0L3 A0A2W1BJJ0 A0A1E1WL08 A0A2A4JDV8 A0A3S2LB02 A0A2A4JDX0 A0A2A4JCM6 A0A2A4JCW2 A0A1E1WU33 A0A2A4JD29 A0A2H1WAI4 A0A2A4JCB8 A0A2A4JDU6 A0A0L7LNR9 A0A0N1IPP3 A0A194QD90 A0A0N1PHM5 A0A194Q7Q6 A0A194Q7A5 A0A0N1I9B8 A0A1E1WV54 A0A1E1WR59 A0A385GMR9 A0A1J1HIE9 U4TRU6 N6UJ02 U4UEL5 B3MWM8 B4NER2 A0A1W4UE05 Q9VYW8 A0A0A1XLB8 B4M238 A0A1Y1NAC9 U5ET31 A0A067QLU2 B4PXW6 B3NUB4 A0A1Y1LJQ0 A0A1J1HKX2 A0A2Z6FLE8 B4H7D8 A0A0K8TRH4 A0A1Q3G4W6 A0A3S2TJN1 Q29G00 Q7KV34 A0A1Q3G4V1 A0A212EH39 D6WTH0 A0A139WE97 A0A212EL62 A0A2P8YX69 B4IK36 A0A182VYD7 A0A212EPE1 A0A1W4WLY4 A0A0N1PEQ1 A0A2S2PG70 F6J1N0 A0A212FGC4 F6JLX3 H9JWK5 A9YGE4 A9YGD5 B0XJU1 F6JGA3
Pubmed
EMBL
BABH01032944
BABH01032945
JTDY01000475
KOB77012.1
KOB77008.1
NWSH01001948
+ More
PCG69594.1 ODYU01012436 SOQ58737.1 SOQ58738.1 PCG69591.1 ODYU01000547 SOQ35490.1 NWSH01000286 PCG77757.1 NWSH01000603 PCG75393.1 ODYU01007302 SOQ49967.1 PCG77758.1 KZ149954 PZC76517.1 PCG69595.1 NWSH01000499 PCG76012.1 SOQ58736.1 KZ150144 PZC72910.1 GDQN01003372 JAT87682.1 PCG69592.1 RSAL01000059 RVE49695.1 PCG69593.1 PCG69599.1 PCG69596.1 GDQN01000534 JAT90520.1 PCG69598.1 SOQ49966.1 PCG69601.1 PCG69600.1 KOB77009.1 KQ460144 KPJ17456.1 KQ459337 KPJ01421.1 KPJ17454.1 KPJ01424.1 KPJ01422.1 KPJ17455.1 GDQN01000227 JAT90827.1 GDQN01001592 JAT89462.1 MG262375 AXX71242.1 CVRI01000006 CRK87815.1 KB631265 ERL84199.1 APGK01033064 KB740860 ENN78592.1 KB632287 ERL91477.1 CH902625 EDV35013.1 CH964239 EDW82231.1 AY061041 AAL28589.1 GBXI01017142 GBXI01002969 JAC97149.1 JAD11323.1 CH940651 EDW65742.1 GEZM01013473 JAV92537.1 GANO01004556 JAB55315.1 KK853171 KDR10313.1 CM000162 EDX01952.2 CH954180 EDV46029.2 GEZM01054023 GEZM01054022 JAV73879.1 CVRI01000010 CRK88711.1 LC384845 BBE28988.1 CH479218 EDW33749.1 GDAI01000636 JAI16967.1 GFDL01000193 JAV34852.1 RSAL01000092 RVE47952.1 CH379065 EAL31429.3 AE014298 AAS65318.1 GFDL01000217 JAV34828.1 AGBW02014964 OWR40805.1 KQ971352 EFA06723.1 KQ971354 KYB26266.1 AGBW02014137 OWR42208.1 PYGN01000305 PSN48850.1 CH480851 EDW51408.1 AGBW02013474 OWR43365.1 KQ459449 KPJ00882.1 GGMR01015810 MBY28429.1 GQ306761 ADG48597.1 AGBW02008687 OWR52782.1 GQ321992 GQ321998 GQ322000 GQ322002 GQ322007 GQ322008 GQ322011 GQ322012 GQ322016 ADG48612.1 ADG48618.1 ADG48620.1 ADG48622.1 ADG48627.1 ADG48628.1 ADG48631.1 ADG48632.1 ADG48636.1 BABH01018451 EU216823 ABW91780.1 EU216814 EU216815 EU216816 EU216818 EU216819 EU216820 EU216821 EU216824 GQ306762 GQ306764 GQ306766 GQ306767 GQ306769 GQ306770 GQ306771 GQ306772 GQ306773 ABW91771.1 ABW91772.1 ABW91773.1 ABW91775.1 ABW91776.1 ABW91777.1 ABW91778.1 ABW91781.1 ADG48598.1 ADG48600.1 ADG48602.1 ADG48603.1 ADG48605.1 ADG48606.1 ADG48607.1 ADG48608.1 ADG48609.1 DS233615 EDS30786.1 GQ317424 ADG48588.1
PCG69594.1 ODYU01012436 SOQ58737.1 SOQ58738.1 PCG69591.1 ODYU01000547 SOQ35490.1 NWSH01000286 PCG77757.1 NWSH01000603 PCG75393.1 ODYU01007302 SOQ49967.1 PCG77758.1 KZ149954 PZC76517.1 PCG69595.1 NWSH01000499 PCG76012.1 SOQ58736.1 KZ150144 PZC72910.1 GDQN01003372 JAT87682.1 PCG69592.1 RSAL01000059 RVE49695.1 PCG69593.1 PCG69599.1 PCG69596.1 GDQN01000534 JAT90520.1 PCG69598.1 SOQ49966.1 PCG69601.1 PCG69600.1 KOB77009.1 KQ460144 KPJ17456.1 KQ459337 KPJ01421.1 KPJ17454.1 KPJ01424.1 KPJ01422.1 KPJ17455.1 GDQN01000227 JAT90827.1 GDQN01001592 JAT89462.1 MG262375 AXX71242.1 CVRI01000006 CRK87815.1 KB631265 ERL84199.1 APGK01033064 KB740860 ENN78592.1 KB632287 ERL91477.1 CH902625 EDV35013.1 CH964239 EDW82231.1 AY061041 AAL28589.1 GBXI01017142 GBXI01002969 JAC97149.1 JAD11323.1 CH940651 EDW65742.1 GEZM01013473 JAV92537.1 GANO01004556 JAB55315.1 KK853171 KDR10313.1 CM000162 EDX01952.2 CH954180 EDV46029.2 GEZM01054023 GEZM01054022 JAV73879.1 CVRI01000010 CRK88711.1 LC384845 BBE28988.1 CH479218 EDW33749.1 GDAI01000636 JAI16967.1 GFDL01000193 JAV34852.1 RSAL01000092 RVE47952.1 CH379065 EAL31429.3 AE014298 AAS65318.1 GFDL01000217 JAV34828.1 AGBW02014964 OWR40805.1 KQ971352 EFA06723.1 KQ971354 KYB26266.1 AGBW02014137 OWR42208.1 PYGN01000305 PSN48850.1 CH480851 EDW51408.1 AGBW02013474 OWR43365.1 KQ459449 KPJ00882.1 GGMR01015810 MBY28429.1 GQ306761 ADG48597.1 AGBW02008687 OWR52782.1 GQ321992 GQ321998 GQ322000 GQ322002 GQ322007 GQ322008 GQ322011 GQ322012 GQ322016 ADG48612.1 ADG48618.1 ADG48620.1 ADG48622.1 ADG48627.1 ADG48628.1 ADG48631.1 ADG48632.1 ADG48636.1 BABH01018451 EU216823 ABW91780.1 EU216814 EU216815 EU216816 EU216818 EU216819 EU216820 EU216821 EU216824 GQ306762 GQ306764 GQ306766 GQ306767 GQ306769 GQ306770 GQ306771 GQ306772 GQ306773 ABW91771.1 ABW91772.1 ABW91773.1 ABW91775.1 ABW91776.1 ABW91777.1 ABW91778.1 ABW91781.1 ADG48598.1 ADG48600.1 ADG48602.1 ADG48603.1 ADG48605.1 ADG48606.1 ADG48607.1 ADG48608.1 ADG48609.1 DS233615 EDS30786.1 GQ317424 ADG48588.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000283053
UP000053240
UP000053268
+ More
UP000183832 UP000030742 UP000019118 UP000007801 UP000007798 UP000192221 UP000008792 UP000027135 UP000002282 UP000008711 UP000008744 UP000001819 UP000000803 UP000007151 UP000007266 UP000245037 UP000001292 UP000075920 UP000192223 UP000002320
UP000183832 UP000030742 UP000019118 UP000007801 UP000007798 UP000192221 UP000008792 UP000027135 UP000002282 UP000008711 UP000008744 UP000001819 UP000000803 UP000007151 UP000007266 UP000245037 UP000001292 UP000075920 UP000192223 UP000002320
Interpro
ProteinModelPortal
H9JIP2
A0A0L7LNI5
A0A0L7LN80
A0A2A4JDG0
A0A2H1X0D0
A0A2H1X0F4
+ More
A0A2A4JCA9 A0A2H1V3S2 A0A2A4K0J1 A0A2A4JV80 A0A2H1WA95 A0A2A4K1F1 A0A2W1BTI3 A0A2A4JBZ6 A0A2A4JX92 A0A2H1X0L3 A0A2W1BJJ0 A0A1E1WL08 A0A2A4JDV8 A0A3S2LB02 A0A2A4JDX0 A0A2A4JCM6 A0A2A4JCW2 A0A1E1WU33 A0A2A4JD29 A0A2H1WAI4 A0A2A4JCB8 A0A2A4JDU6 A0A0L7LNR9 A0A0N1IPP3 A0A194QD90 A0A0N1PHM5 A0A194Q7Q6 A0A194Q7A5 A0A0N1I9B8 A0A1E1WV54 A0A1E1WR59 A0A385GMR9 A0A1J1HIE9 U4TRU6 N6UJ02 U4UEL5 B3MWM8 B4NER2 A0A1W4UE05 Q9VYW8 A0A0A1XLB8 B4M238 A0A1Y1NAC9 U5ET31 A0A067QLU2 B4PXW6 B3NUB4 A0A1Y1LJQ0 A0A1J1HKX2 A0A2Z6FLE8 B4H7D8 A0A0K8TRH4 A0A1Q3G4W6 A0A3S2TJN1 Q29G00 Q7KV34 A0A1Q3G4V1 A0A212EH39 D6WTH0 A0A139WE97 A0A212EL62 A0A2P8YX69 B4IK36 A0A182VYD7 A0A212EPE1 A0A1W4WLY4 A0A0N1PEQ1 A0A2S2PG70 F6J1N0 A0A212FGC4 F6JLX3 H9JWK5 A9YGE4 A9YGD5 B0XJU1 F6JGA3
A0A2A4JCA9 A0A2H1V3S2 A0A2A4K0J1 A0A2A4JV80 A0A2H1WA95 A0A2A4K1F1 A0A2W1BTI3 A0A2A4JBZ6 A0A2A4JX92 A0A2H1X0L3 A0A2W1BJJ0 A0A1E1WL08 A0A2A4JDV8 A0A3S2LB02 A0A2A4JDX0 A0A2A4JCM6 A0A2A4JCW2 A0A1E1WU33 A0A2A4JD29 A0A2H1WAI4 A0A2A4JCB8 A0A2A4JDU6 A0A0L7LNR9 A0A0N1IPP3 A0A194QD90 A0A0N1PHM5 A0A194Q7Q6 A0A194Q7A5 A0A0N1I9B8 A0A1E1WV54 A0A1E1WR59 A0A385GMR9 A0A1J1HIE9 U4TRU6 N6UJ02 U4UEL5 B3MWM8 B4NER2 A0A1W4UE05 Q9VYW8 A0A0A1XLB8 B4M238 A0A1Y1NAC9 U5ET31 A0A067QLU2 B4PXW6 B3NUB4 A0A1Y1LJQ0 A0A1J1HKX2 A0A2Z6FLE8 B4H7D8 A0A0K8TRH4 A0A1Q3G4W6 A0A3S2TJN1 Q29G00 Q7KV34 A0A1Q3G4V1 A0A212EH39 D6WTH0 A0A139WE97 A0A212EL62 A0A2P8YX69 B4IK36 A0A182VYD7 A0A212EPE1 A0A1W4WLY4 A0A0N1PEQ1 A0A2S2PG70 F6J1N0 A0A212FGC4 F6JLX3 H9JWK5 A9YGE4 A9YGD5 B0XJU1 F6JGA3
Ontologies
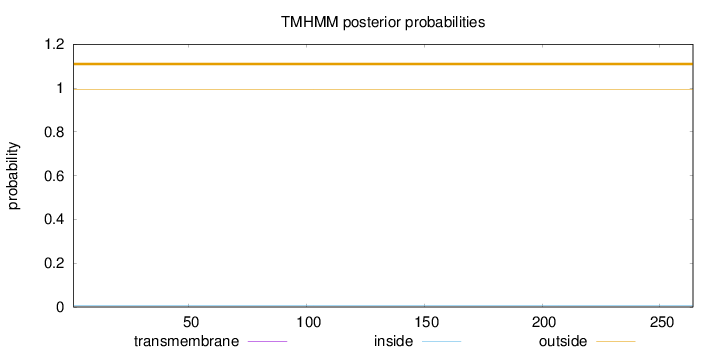
Topology
Length:
264
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00041
Exp number, first 60 AAs:
0.00041
Total prob of N-in:
0.00589
outside
1 - 264
Population Genetic Test Statistics
Pi
127.837546
Theta
131.087249
Tajima's D
-0.304305
CLR
0.830289
CSRT
0.286135693215339
Interpretation
Uncertain
