Gene
KWMTBOMO08466
Pre Gene Modal
BGIBMGA009392
Annotation
Ecdysteroid_22-kinase_[Operophtera_brumata]
Location in the cell
Cytoplasmic Reliability : 1.964
Sequence
CDS
ATGGGTGAAAACCTCGTCGAATCAGTATTGATCGACTATCAAATCTCCAAAGAAACGAGTCCCGTCTGCGACATCCAATACATGATCTTCAATTGCACCGACCACAAAACCAGACTCAAATATTTTAACGAATGGCTCGACTATTATCACGCAGAATTAGAAAAACGCCTAACGAATTTCGGACTTAAAATCGAAAATGTCTACCCCAGGGAACAATTTGACGCCGACATGAAGAAATATAGTGGAATGATGTTGGGACTCTCTACTATTCTCGCAAGCGTACTAACCATGAATTCAGAGGACGCTGGGAAAATGAAGGAAACGATGGAATCGACATCACCAGATGAACTACCAGCAAACAACGAAGTAGAAGATTACAATGTTGAGCACACGATAAAATTTAAAACTAGACTCGAAGGTCTTATAGATAGTTATCTTGAATTCAATTTAATTTAG
Protein
MGENLVESVLIDYQISKETSPVCDIQYMIFNCTDHKTRLKYFNEWLDYYHAELEKRLTNFGLKIENVYPREQFDADMKKYSGMMLGLSTILASVLTMNSEDAGKMKETMESTSPDELPANNEVEDYNVEHTIKFKTRLEGLIDSYLEFNLI
Summary
Uniprot
H9JIP2
A0A0L7L0L9
A0A0L7LN80
A0A0L7LNK2
A0A0L7LNI5
A0A2H1V3S2
+ More
A0A2A4JDG0 A0A2H1WA95 A0A2A4JDX0 A0A2H1X0D0 A0A2H1X0F4 A0A0L7LNR9 A0A2A4JBZ6 A0A2W1BTI3 A0A2A4K1F1 A0A2A4JCA9 A0A1E1W2T0 A0A2A4K0J1 A0A3S2LB02 A0A2A4JV80 A0A2W1BJJ0 A0A1E1WU33 A0A2A4JCB8 A0A1E1WL08 A0A2H1X0L3 A0A2A4JDU6 A0A2H1WAI4 A0A2A4JCW2 A0A2A4JDV8 A0A194QD90 A0A2A4JD29 A0A0N1IPP3 A0A2A4JX92 A0A0N1I9B8 A0A2A4JCM6 A0A194Q7Q6 A0A194Q7A5 A0A212F2J2 A0A0L7LUY7 A0A2A4JDZ5 Q9VW98 A0A2A4JF80 A0A0N1I9M5 B4NUB4 A0A0L7K3X1 A0A1I8P5D0 A0A182M5W7 B4IA62 A0A182T270 A0A2W1BGZ2 B4QRV2 A0A2C9GQW4 A0A0N1PGW5 A0A0S1MMB6 A0A182L9J1 A0A0A1XLB8 A0A182UWA2 A0A182JA39 A0A2H1WHE7 A0A182R4V1 A0A182YAE9 A0A2W1BSP2 B3M4Q7 F5HML7 A0A0N1IEA0 A0A182PGU5 A0A182VWJ2 A0A0L0BW33 A0A0L7KXT2 A0A2C9GQM5 A0A2H1V9H4 A0A182L9J8 F5HML8 Q17DF4 A0A182T410 B4PFD0 A0A1A9X5H2 A0A2H1V9I6 Q9VYW8 A0A182UNS4 A0A2H1VEU0 A0A1W4W5N2 A0A2W1BG42 B3NE52 A0A1S4F7C6 B4M238 A0A437BPI8 B4MKV4 A0A2W1BMZ2 A0A0N1PEQ1 A0A1L8DN51 A0A2H1V9N0 A0A0K8TRH4 H9JWQ7 A0A182TQB9 A0A0N0PB04
A0A2A4JDG0 A0A2H1WA95 A0A2A4JDX0 A0A2H1X0D0 A0A2H1X0F4 A0A0L7LNR9 A0A2A4JBZ6 A0A2W1BTI3 A0A2A4K1F1 A0A2A4JCA9 A0A1E1W2T0 A0A2A4K0J1 A0A3S2LB02 A0A2A4JV80 A0A2W1BJJ0 A0A1E1WU33 A0A2A4JCB8 A0A1E1WL08 A0A2H1X0L3 A0A2A4JDU6 A0A2H1WAI4 A0A2A4JCW2 A0A2A4JDV8 A0A194QD90 A0A2A4JD29 A0A0N1IPP3 A0A2A4JX92 A0A0N1I9B8 A0A2A4JCM6 A0A194Q7Q6 A0A194Q7A5 A0A212F2J2 A0A0L7LUY7 A0A2A4JDZ5 Q9VW98 A0A2A4JF80 A0A0N1I9M5 B4NUB4 A0A0L7K3X1 A0A1I8P5D0 A0A182M5W7 B4IA62 A0A182T270 A0A2W1BGZ2 B4QRV2 A0A2C9GQW4 A0A0N1PGW5 A0A0S1MMB6 A0A182L9J1 A0A0A1XLB8 A0A182UWA2 A0A182JA39 A0A2H1WHE7 A0A182R4V1 A0A182YAE9 A0A2W1BSP2 B3M4Q7 F5HML7 A0A0N1IEA0 A0A182PGU5 A0A182VWJ2 A0A0L0BW33 A0A0L7KXT2 A0A2C9GQM5 A0A2H1V9H4 A0A182L9J8 F5HML8 Q17DF4 A0A182T410 B4PFD0 A0A1A9X5H2 A0A2H1V9I6 Q9VYW8 A0A182UNS4 A0A2H1VEU0 A0A1W4W5N2 A0A2W1BG42 B3NE52 A0A1S4F7C6 B4M238 A0A437BPI8 B4MKV4 A0A2W1BMZ2 A0A0N1PEQ1 A0A1L8DN51 A0A2H1V9N0 A0A0K8TRH4 H9JWQ7 A0A182TQB9 A0A0N0PB04
Pubmed
EMBL
BABH01032944
BABH01032945
JTDY01003763
KOB69043.1
JTDY01000475
KOB77008.1
+ More
KOB77010.1 KOB77012.1 ODYU01000547 SOQ35490.1 NWSH01001948 PCG69594.1 ODYU01007302 SOQ49967.1 PCG69593.1 ODYU01012436 SOQ58737.1 SOQ58738.1 KOB77009.1 PCG69595.1 KZ149954 PZC76517.1 NWSH01000286 PCG77758.1 PCG69591.1 GDQN01009843 JAT81211.1 PCG77757.1 RSAL01000059 RVE49695.1 NWSH01000603 PCG75393.1 KZ150144 PZC72910.1 GDQN01000534 JAT90520.1 PCG69601.1 GDQN01003372 JAT87682.1 SOQ58736.1 PCG69600.1 SOQ49966.1 PCG69596.1 PCG69592.1 KQ459337 KPJ01421.1 PCG69598.1 KQ460144 KPJ17456.1 NWSH01000499 PCG76012.1 KPJ17455.1 PCG69599.1 KPJ01424.1 KPJ01422.1 AGBW02010741 OWR47943.1 JTDY01000032 KOB79293.1 NWSH01001934 PCG69632.1 AE014296 BT022472 AAF49050.1 AAY54888.1 NWSH01001667 PCG70469.1 KQ459449 KPJ00881.1 CH983834 EDX16561.1 JTDY01011879 KOB52369.1 AXCM01013491 AXCM01013492 CH480826 EDW44175.1 KZ150068 PZC74088.1 CM000363 CM002912 EDX11198.1 KMZ00708.1 APCN01000881 KQ461184 KPJ07528.1 KR821066 ALL42054.1 GBXI01017142 GBXI01002969 JAC97149.1 JAD11323.1 ODYU01008699 SOQ52500.1 KZ149965 PZC76227.1 CH902618 EDV39456.1 AAAB01008987 EGK97539.1 KQ458721 KPJ05409.1 JRES01001250 KNC24233.1 JTDY01004714 KOB67844.1 ODYU01001386 SOQ37500.1 EGK97540.1 CH477296 EAT44383.1 CM000159 EDW95216.1 SOQ37495.1 AY061041 AAL28589.1 ODYU01002157 SOQ39296.1 PZC74089.1 CH954178 EDV52476.1 CH940651 EDW65742.1 RSAL01000022 RVE52328.1 CH963847 EDW72879.2 PZC74090.1 KPJ00882.1 GFDF01006277 JAV07807.1 SOQ37496.1 GDAI01000636 JAI16967.1 BABH01018468 KPJ07527.1
KOB77010.1 KOB77012.1 ODYU01000547 SOQ35490.1 NWSH01001948 PCG69594.1 ODYU01007302 SOQ49967.1 PCG69593.1 ODYU01012436 SOQ58737.1 SOQ58738.1 KOB77009.1 PCG69595.1 KZ149954 PZC76517.1 NWSH01000286 PCG77758.1 PCG69591.1 GDQN01009843 JAT81211.1 PCG77757.1 RSAL01000059 RVE49695.1 NWSH01000603 PCG75393.1 KZ150144 PZC72910.1 GDQN01000534 JAT90520.1 PCG69601.1 GDQN01003372 JAT87682.1 SOQ58736.1 PCG69600.1 SOQ49966.1 PCG69596.1 PCG69592.1 KQ459337 KPJ01421.1 PCG69598.1 KQ460144 KPJ17456.1 NWSH01000499 PCG76012.1 KPJ17455.1 PCG69599.1 KPJ01424.1 KPJ01422.1 AGBW02010741 OWR47943.1 JTDY01000032 KOB79293.1 NWSH01001934 PCG69632.1 AE014296 BT022472 AAF49050.1 AAY54888.1 NWSH01001667 PCG70469.1 KQ459449 KPJ00881.1 CH983834 EDX16561.1 JTDY01011879 KOB52369.1 AXCM01013491 AXCM01013492 CH480826 EDW44175.1 KZ150068 PZC74088.1 CM000363 CM002912 EDX11198.1 KMZ00708.1 APCN01000881 KQ461184 KPJ07528.1 KR821066 ALL42054.1 GBXI01017142 GBXI01002969 JAC97149.1 JAD11323.1 ODYU01008699 SOQ52500.1 KZ149965 PZC76227.1 CH902618 EDV39456.1 AAAB01008987 EGK97539.1 KQ458721 KPJ05409.1 JRES01001250 KNC24233.1 JTDY01004714 KOB67844.1 ODYU01001386 SOQ37500.1 EGK97540.1 CH477296 EAT44383.1 CM000159 EDW95216.1 SOQ37495.1 AY061041 AAL28589.1 ODYU01002157 SOQ39296.1 PZC74089.1 CH954178 EDV52476.1 CH940651 EDW65742.1 RSAL01000022 RVE52328.1 CH963847 EDW72879.2 PZC74090.1 KPJ00882.1 GFDF01006277 JAV07807.1 SOQ37496.1 GDAI01000636 JAI16967.1 BABH01018468 KPJ07527.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000283053
UP000053268
UP000053240
+ More
UP000007151 UP000000803 UP000000304 UP000095300 UP000075883 UP000001292 UP000075901 UP000075840 UP000075882 UP000075903 UP000075880 UP000075900 UP000076408 UP000007801 UP000007062 UP000075885 UP000075920 UP000037069 UP000008820 UP000002282 UP000091820 UP000192221 UP000008711 UP000008792 UP000007798 UP000075902
UP000007151 UP000000803 UP000000304 UP000095300 UP000075883 UP000001292 UP000075901 UP000075840 UP000075882 UP000075903 UP000075880 UP000075900 UP000076408 UP000007801 UP000007062 UP000075885 UP000075920 UP000037069 UP000008820 UP000002282 UP000091820 UP000192221 UP000008711 UP000008792 UP000007798 UP000075902
Pfam
PF02958 EcKinase
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
H9JIP2
A0A0L7L0L9
A0A0L7LN80
A0A0L7LNK2
A0A0L7LNI5
A0A2H1V3S2
+ More
A0A2A4JDG0 A0A2H1WA95 A0A2A4JDX0 A0A2H1X0D0 A0A2H1X0F4 A0A0L7LNR9 A0A2A4JBZ6 A0A2W1BTI3 A0A2A4K1F1 A0A2A4JCA9 A0A1E1W2T0 A0A2A4K0J1 A0A3S2LB02 A0A2A4JV80 A0A2W1BJJ0 A0A1E1WU33 A0A2A4JCB8 A0A1E1WL08 A0A2H1X0L3 A0A2A4JDU6 A0A2H1WAI4 A0A2A4JCW2 A0A2A4JDV8 A0A194QD90 A0A2A4JD29 A0A0N1IPP3 A0A2A4JX92 A0A0N1I9B8 A0A2A4JCM6 A0A194Q7Q6 A0A194Q7A5 A0A212F2J2 A0A0L7LUY7 A0A2A4JDZ5 Q9VW98 A0A2A4JF80 A0A0N1I9M5 B4NUB4 A0A0L7K3X1 A0A1I8P5D0 A0A182M5W7 B4IA62 A0A182T270 A0A2W1BGZ2 B4QRV2 A0A2C9GQW4 A0A0N1PGW5 A0A0S1MMB6 A0A182L9J1 A0A0A1XLB8 A0A182UWA2 A0A182JA39 A0A2H1WHE7 A0A182R4V1 A0A182YAE9 A0A2W1BSP2 B3M4Q7 F5HML7 A0A0N1IEA0 A0A182PGU5 A0A182VWJ2 A0A0L0BW33 A0A0L7KXT2 A0A2C9GQM5 A0A2H1V9H4 A0A182L9J8 F5HML8 Q17DF4 A0A182T410 B4PFD0 A0A1A9X5H2 A0A2H1V9I6 Q9VYW8 A0A182UNS4 A0A2H1VEU0 A0A1W4W5N2 A0A2W1BG42 B3NE52 A0A1S4F7C6 B4M238 A0A437BPI8 B4MKV4 A0A2W1BMZ2 A0A0N1PEQ1 A0A1L8DN51 A0A2H1V9N0 A0A0K8TRH4 H9JWQ7 A0A182TQB9 A0A0N0PB04
A0A2A4JDG0 A0A2H1WA95 A0A2A4JDX0 A0A2H1X0D0 A0A2H1X0F4 A0A0L7LNR9 A0A2A4JBZ6 A0A2W1BTI3 A0A2A4K1F1 A0A2A4JCA9 A0A1E1W2T0 A0A2A4K0J1 A0A3S2LB02 A0A2A4JV80 A0A2W1BJJ0 A0A1E1WU33 A0A2A4JCB8 A0A1E1WL08 A0A2H1X0L3 A0A2A4JDU6 A0A2H1WAI4 A0A2A4JCW2 A0A2A4JDV8 A0A194QD90 A0A2A4JD29 A0A0N1IPP3 A0A2A4JX92 A0A0N1I9B8 A0A2A4JCM6 A0A194Q7Q6 A0A194Q7A5 A0A212F2J2 A0A0L7LUY7 A0A2A4JDZ5 Q9VW98 A0A2A4JF80 A0A0N1I9M5 B4NUB4 A0A0L7K3X1 A0A1I8P5D0 A0A182M5W7 B4IA62 A0A182T270 A0A2W1BGZ2 B4QRV2 A0A2C9GQW4 A0A0N1PGW5 A0A0S1MMB6 A0A182L9J1 A0A0A1XLB8 A0A182UWA2 A0A182JA39 A0A2H1WHE7 A0A182R4V1 A0A182YAE9 A0A2W1BSP2 B3M4Q7 F5HML7 A0A0N1IEA0 A0A182PGU5 A0A182VWJ2 A0A0L0BW33 A0A0L7KXT2 A0A2C9GQM5 A0A2H1V9H4 A0A182L9J8 F5HML8 Q17DF4 A0A182T410 B4PFD0 A0A1A9X5H2 A0A2H1V9I6 Q9VYW8 A0A182UNS4 A0A2H1VEU0 A0A1W4W5N2 A0A2W1BG42 B3NE52 A0A1S4F7C6 B4M238 A0A437BPI8 B4MKV4 A0A2W1BMZ2 A0A0N1PEQ1 A0A1L8DN51 A0A2H1V9N0 A0A0K8TRH4 H9JWQ7 A0A182TQB9 A0A0N0PB04
Ontologies
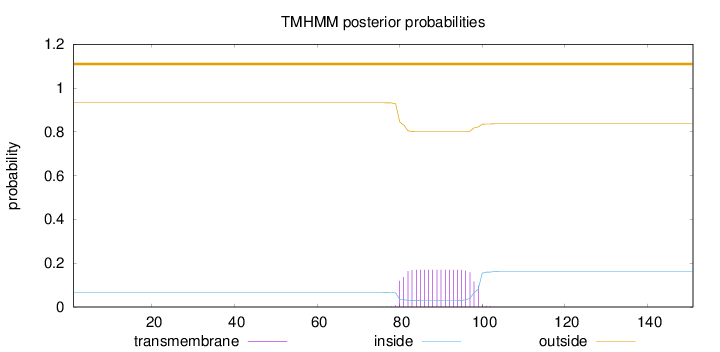
Topology
Length:
151
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.19668
Exp number, first 60 AAs:
0.00062
Total prob of N-in:
0.06697
outside
1 - 151
Population Genetic Test Statistics
Pi
279.697919
Theta
200.2539
Tajima's D
1.185299
CLR
0
CSRT
0.703014849257537
Interpretation
Uncertain
