Gene
KWMTBOMO08465 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009423
Annotation
PREDICTED:_uncharacterized_protein_LOC105841524_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.076
Sequence
CDS
ATGGAATTTCTGATAATATTTGTGGGGGTCCTCCTCGCGGCATCAGCGACCGCTGATCAAGATGTGGCTGAAGATGTTCCCAGTCGCAGATATTTACAAATTATTGAACAAAACGTTAAAAGACCGAATCCTTACGAGATTGTGAAGCCGTGGCCGTACGGAAGTCAACGCGATGTCGTCTCTAAATCAGTGAGGGTAGATTACAATCCGCTAGATACTAATGTCAACTCTAATGTAAACGGAAACAATGCTATGAAGATCGCAAATCTCATGTCAGTTATGGCCAAAAACTCGGTTGCAGATCAAAATGCAGGACGCAATGGTTTTGGCCAAGAAGTAAACTCACAAATTCGTATGTATCATGTTCCAAGCTCTGAAAATCAAAATTCGTTCAGTGTTGTACTCAAAAAAAGAATGTCACCCGAAGGAGAAATTGAAACCGTTGAATACAAAGTACCGGACCAATATATCCAGCCCCAGGATCTCCGAAGCAATAGTTTGAATAATGTTCAGTCCCTATCGAGTTACAAGATAACAGATACTTTAAGAGAACCAAACAAGTTTGTCGCAGAACAAGAGTTTGCCTCCAGTAAGTACATGCATAAAGCACCGAACACGTATATTGATTCTCAGTTAAAGCTCGGAAGCAATACGCATGATGAAATACGTGACGAATCAACTTCGGGATCGGTAGAATACAAAGAAGACGCTGAATTAAATTATATAAGAGCCGAAAATCTGCGTGGCGGTGCTTACAGTGATGAATACGCAAAAGAAAAGAGGTTCTATGAATCACGCAGTAATAGTTTGAACCACGATCAATATAAAGCTATAAATCAGGAAGCAGGATACAACATTGATGGACAAAGGCAGTTGGATCCTACATTTTTAGAATCTGATATTGAAGTGGTTAGTGAAAAAAAAGAAAATTATAGTCCATATGACTTATACGATAAACGAGAGAACGTGATAAAAGATAGGTATACCTATGGTCACAGTCCTGCTCAACGTTATCAAAACGCTTTTGCAGTTAATAATTACGGCTCTGGAAGGAATAAAGTGAAACCGTACTACGGAAATTACTTCGAAAACTATCAACAGCCACAAATCGTAGATACCACGCAAAGTATGAAGGAAAAAATAGATTTCAATACTGAGAGCGTCAGTCCAGACTTTGAATATAAAAGCAAAATGAAAAACTATCAAAACTATGTAGACGTTCAGCAAAAACCGTACGGTAATAATGAACTTGATAGGTTTAAAAGCACTGCAGGATATACCCATTACGAAAATCCATTAACCAGAGTTGATAAAAAACAAAACTATATTAATTACGAACAACAATATTACCCAACCAGTTCATCTGTTCGAGACAGCAAGGGTTCGATAAAATTAACTGAGTTAAAAAAAAACAATGTTAATTATCAAGCCGGCACAGTGCTCCAGGAGTTCCAAGATACAGAGGAATTTAAAAACCAGTACAATTCTAAAGTAAATTTTGTTGCACCAGTTGATAAATGGGGCCAAACTAAAGAAGAAACCCTTAGTGTTTTAGAACGTGGAATATTAATAGATCTTATAGCGCAAAGTATAAATAAATTTAGGATTACACTGAAGGAACTTCAAGTCGCAGGTGCTCGTTTTGGAAATATGCAGTGCGCTGAAAATGGTTCGCTCTTACGACGAGCTGCAGCGACGAAATTAGCACAGGATGATTCGAATTCAAATTCGCTGAAGTTCAAAGCCAGACAAACAGAAAAAGTGAAATCGAAATTAAATATTCCCACTATCTTTGGAAATGAACAGATAAAAACAGAAAACGAAGATGACGTAGAGACTCAGTTTTCAGTAAAGGAGAAATTCAGTCCAAACATGAATAATAATAATCAATTTAATTACATGGCAAGCGATAAAAAGTTAGAATTGATGCAAAGTCAACAATTACCGTTGAACTATCAAGAATTTATCATCGGAGAACAGTCAAGATTTGAGCCAGCGCACCTTAAAGAAGAACAAGGCTCATATCAATTTACTTCAGTGAATAATCCAACTGGCTATCAAACTGGCTTGTATCAAGGAAACAAAATTGATTTTACTTCATTAGGCGAACAGAGGATTATCGACCAAAGAGTTCCTTATAATCGTGAAGTCATCGAAAATAGCTCCAATGAAAGATTTAACGAAGAACATAAGATGTCGATAAGCGGGGAATTAAGCGGCTCTAACCAAAATATGCCGGCAAAAAATTACCAGAATTTGAGGTATAAAGCTCAAATTAAAGACGATTACGATCGAAGGGACAGTCAAACCATTAAGAATTCCATCTTCATTAACAATAAATATAATTTAAACAACGTCAATGATCGATTTTCAATTGCTAGTATACTAAAAGATTCTATGGGGAGCTTATCCGATGAAGAAATATTCGTGATGCAGCTCAACTCGATCGACCAGAACAACAATTTGCCACTGCTGAGGGAGAAACTCATCGAACTCCTTAAAATGATTAGATCGAAATGGACGTATCTACAGAAATTCATAAGCATAATTTTGAAACTGGACAATGGAAACTGTTTCGTGAACAGCGATCAGTTAAAGGCCTTGTTTAATAATCAAGCTTATAGTGATTTGGAAATGACGAAAGTAGGTTACGGAAATGCTTACATGAAGTATTATCCGTGTAGAATCGGCACTCAAATGATAACTCCTTGTGATCGTTGTTTCTGCAAATACAATGGAATCTTGGTCTGTCAGAAAAAAGTACTTTGTGGACGCTGA
Protein
MEFLIIFVGVLLAASATADQDVAEDVPSRRYLQIIEQNVKRPNPYEIVKPWPYGSQRDVVSKSVRVDYNPLDTNVNSNVNGNNAMKIANLMSVMAKNSVADQNAGRNGFGQEVNSQIRMYHVPSSENQNSFSVVLKKRMSPEGEIETVEYKVPDQYIQPQDLRSNSLNNVQSLSSYKITDTLREPNKFVAEQEFASSKYMHKAPNTYIDSQLKLGSNTHDEIRDESTSGSVEYKEDAELNYIRAENLRGGAYSDEYAKEKRFYESRSNSLNHDQYKAINQEAGYNIDGQRQLDPTFLESDIEVVSEKKENYSPYDLYDKRENVIKDRYTYGHSPAQRYQNAFAVNNYGSGRNKVKPYYGNYFENYQQPQIVDTTQSMKEKIDFNTESVSPDFEYKSKMKNYQNYVDVQQKPYGNNELDRFKSTAGYTHYENPLTRVDKKQNYINYEQQYYPTSSSVRDSKGSIKLTELKKNNVNYQAGTVLQEFQDTEEFKNQYNSKVNFVAPVDKWGQTKEETLSVLERGILIDLIAQSINKFRITLKELQVAGARFGNMQCAENGSLLRRAAATKLAQDDSNSNSLKFKARQTEKVKSKLNIPTIFGNEQIKTENEDDVETQFSVKEKFSPNMNNNNQFNYMASDKKLELMQSQQLPLNYQEFIIGEQSRFEPAHLKEEQGSYQFTSVNNPTGYQTGLYQGNKIDFTSLGEQRIIDQRVPYNREVIENSSNERFNEEHKMSISGELSGSNQNMPAKNYQNLRYKAQIKDDYDRRDSQTIKNSIFINNKYNLNNVNDRFSIASILKDSMGSLSDEEIFVMQLNSIDQNNNLPLLREKLIELLKMIRSKWTYLQKFISIILKLDNGNCFVNSDQLKALFNNQAYSDLEMTKVGYGNAYMKYYPCRIGTQMITPCDRCFCKYNGILVCQKKVLCGR
Summary
Uniprot
Pubmed
EMBL
Proteomes
PRIDE
ProteinModelPortal
Ontologies
GO
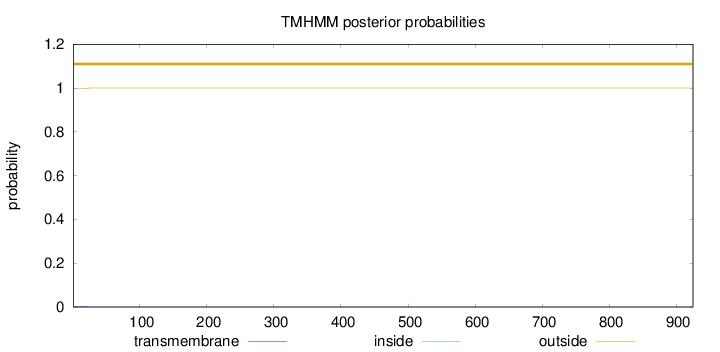
Topology
SignalP
Position: 1 - 18,
Likelihood: 0.998029
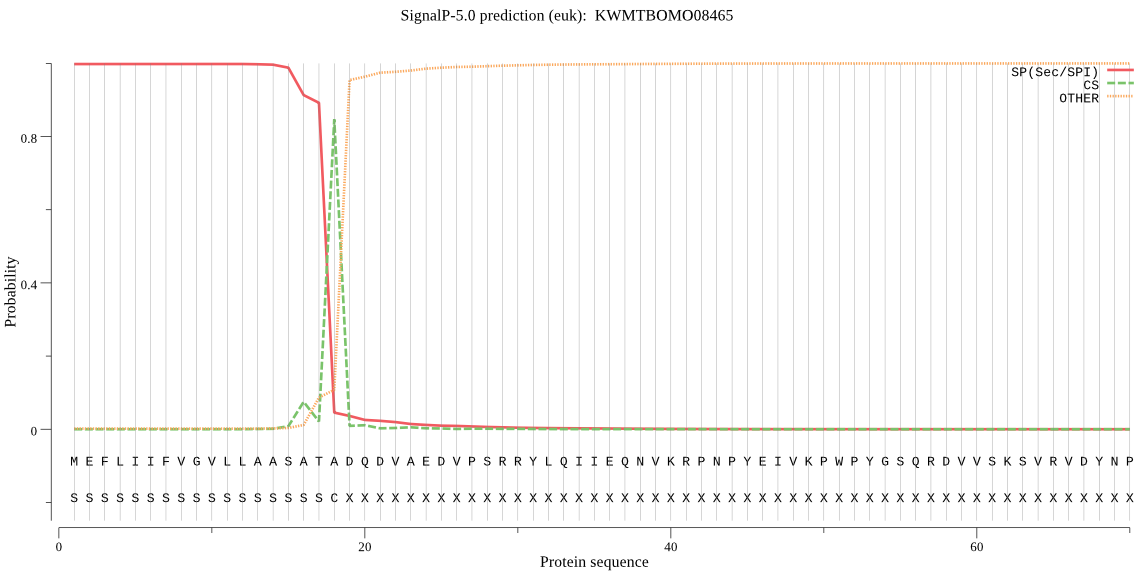
Length:
925
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0503000000000001
Exp number, first 60 AAs:
0.0486
Total prob of N-in:
0.00258
outside
1 - 925
Population Genetic Test Statistics
Pi
245.330218
Theta
193.338756
Tajima's D
0.790212
CLR
0.502692
CSRT
0.600969951502425
Interpretation
Uncertain
