Gene
KWMTBOMO08461 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009420
Annotation
Electron_transfer_flavoprotein-ubiquinone_oxidoreductase?_mitochondrial_[Papilio_xuthus]
Full name
Dihydrolipoyl dehydrogenase
Location in the cell
Cytoplasmic Reliability : 1.276 Mitochondrial Reliability : 1.905
Sequence
CDS
ATGGCGGTGGCTATTGTTAGTTCGGCTCGACAGGTGGGGAGGCTAACAAACGCAGCCAGGAGGCTTTATTCTGATGTATATCCTAAAATATCAACCCATTATACAATACATCCAAGAGATAAGGATCCAAGATGGAAAGAGATAAATATGGAACGCGTAGCGGAGGAGACGGACATCCTCATTATCGGTGGGGGCCCAGCGGGTATGGCAGCTGCAATCCGAGCAAGACAAATAGCAGCTGAAAAGGGGGCAGAAGTAAGAGTGACTTTACTGGAAAAGGCAGCAGAGACTGGAGGGCATATATTGTCAGGAGCCTGTGTTGATCCAATAGCTTTGAATGAGTTAATCCCAGACTGGAAAGAGCGAGGAGCTCCTATGAATACTCCAGTTACATCGGACAAGTTTGGTCTGTTGACGAAGAGTAGCAGAATACCTATACCTGTATTTCCTGGTTTACCGAATTACAATCATGGTAACTACGTTGTGAGACTGGGACATCTAGTTAAATGGCTTGGTGAGCAAGCTGAAGAACTCGGTGCTGAGGTGTGGCCGGGATGCGCTGGAGCCGAGGTGCTGTACCGCGAGGACGGGTCGGTGCGCGGGGTGGCCACGGGGTGTCATGGACATTTAACGAAAATGGTCTCGAACAAGTTCAATCTCAGGGAGAAGAGCGAGCCACAGTCTTATGGTATTGGATTGAAAGAGCTTTGGGAAGTTAAGCCAGAGAACCACAAACCAGGTTTAGTTGAACACACAATAGGTTGGCCATTAGATAAGAACACATACGGGGGATCGTTCATATACCACTTAAATGTGGCAGAGGGCGAATCACCCCTAGTGGCCGTCGGCTTCGTGGTGGGGCTGGACTACACCAACCCTTACATTAGTCCCTTCAGAGAATTCCAGAGATTCAAGTTGCATCCTTATGTTAATCCTGTGTTCGAAGGCGGGTCCCGCATCGCGTACGGCGCGCGGGCGCTGGTGGAGGGCGGCTGGCAGTGCCTGCCCCGGCCGGTGTTCCCGGGGGGCGTGCTGGCCGGGGACACGGCGGGCTTCCTCAACGTGCCCCGGATCAAGGGCACGCACAACGCCATGAAGAGTGGCATGCTAGCCGCCGAGTCCGCCATGGAGCTGGTGCTGTCCGGGGACGCGACACACGATCGCGGAGTCGTTCCGGAGTCTTACGAAGAGAAACTAAAAAATTCATTCGTCTACAAAGAACTCAAGGAGGTTCGCAACTGTCGGCCATCGTTCCACACGAAGCTGGGCCTGTACGGAGGCGTCGCGTACTCCGCCTTCAGCACGCTCGTGCGCGGGAAGGAACCCTGGACCCTCAGCCACGGAGGTGCAGATCACTCACGCCTGCGGCCGGCGCAGGAGTGCCAACCCATCGAGTACCCCAAGCCGGACGGGGTGGTTACCTTCGATCTGCTGTCCTCTGTTGCGCTCACAGGCACAAATCACGAAGCGGACCAGCCTCCGCATCTGACGTTAAAAGACGATACGGTGCCGGTTAAGACCAATCTAGGAGTATACGATGGACCCGAAGCCAGGTTTTGTCCTGCAGGTGTATACGAGTTCGTACCGTTGGAGGAGGGTGATGGTCAGAGGTTGCAGATCAATGCTCAGAACTGCATCCACTGTAAAACGTGCGACATTAAAGACCCGTCGCAGAACATCAACTGGGTTGTGCCCGAGGGCGGCGGCGGACCTGCCTACAACGGAATGTAA
Protein
MAVAIVSSARQVGRLTNAARRLYSDVYPKISTHYTIHPRDKDPRWKEINMERVAEETDILIIGGGPAGMAAAIRARQIAAEKGAEVRVTLLEKAAETGGHILSGACVDPIALNELIPDWKERGAPMNTPVTSDKFGLLTKSSRIPIPVFPGLPNYNHGNYVVRLGHLVKWLGEQAEELGAEVWPGCAGAEVLYREDGSVRGVATGCHGHLTKMVSNKFNLREKSEPQSYGIGLKELWEVKPENHKPGLVEHTIGWPLDKNTYGGSFIYHLNVAEGESPLVAVGFVVGLDYTNPYISPFREFQRFKLHPYVNPVFEGGSRIAYGARALVEGGWQCLPRPVFPGGVLAGDTAGFLNVPRIKGTHNAMKSGMLAAESAMELVLSGDATHDRGVVPESYEEKLKNSFVYKELKEVRNCRPSFHTKLGLYGGVAYSAFSTLVRGKEPWTLSHGGADHSRLRPAQECQPIEYPKPDGVVTFDLLSSVALTGTNHEADQPPHLTLKDDTVPVKTNLGVYDGPEARFCPAGVYEFVPLEEGDGQRLQINAQNCIHCKTCDIKDPSQNINWVVPEGGGGPAYNGM
Summary
Catalytic Activity
(R)-N(6)-dihydrolipoyl-L-lysyl-[protein] + NAD(+) = (R)-N(6)-lipoyl-L-lysyl-[protein] + H(+) + NADH
Cofactor
FAD
Miscellaneous
The active site is a redox-active disulfide bond.
Similarity
Belongs to the class-I pyridine nucleotide-disulfide oxidoreductase family.
Uniprot
H9JIR9
A0A194Q7R1
A0A1Z2RRI8
A0A3S2PF61
A0A212EWT6
A0A0C9RK51
+ More
A0A232FMI1 K7IXE6 A0A2J7PZJ7 J3JTE9 V5GUY2 A0A1B6C0L4 A0A182RUZ2 A0A151XJD1 A0A182NMH1 F4X3U3 A0A182Q739 A0A3L8DXW6 A0A026WNE4 A0A182M6W2 A0A182Y2X4 A0A158P050 A0A0P6J107 D2A0Q9 E2A3X9 A0A195E0P7 A0A195F1N3 A0A067RE05 A0A182PPA5 U5ES10 A0A182FE41 A0A195BFJ9 A0A084W2Q5 A0A2M4BHQ0 A0A1Y1KN64 W5JFW5 A0A182VKC9 A0A182IN15 A0A2M3Z244 A0A182XM95 A0A195CA52 A0A182WMD0 A0A182I230 A0A182GB59 A0A2A3E9F3 A0A2M4A6L1 A0A182TP92 Q7Q6E7 A0A2M4A6I3 A0A0B4J2M5 A0A182JRN1 A0A310SP62 A0A0A9WLX0 B0VZ76 R4WSM6 A0A1Q3FI44 T1DPN1 A0A0T6B555 A0A154NWP3 A0A182KPU7 A0A0L7QW90 J9JTH9 A0A0P4VSJ0 T1HF19 A0A2R5LJQ2 A0A0C9Q061 A0A1L8E0P0 A0A336KS65 A0A336M9M1 A0A224XDK6 A0A0V0G5A3 A0A2S2QCW4 Q171U3 E0VBI1 A0A1W4WSQ7 A0A2M4BIZ2 A0A1W4X2C3 A0A023FAJ2 W8BP19 A7RNA5 A0A1Z5KUI9 B3N6T6 A0A1S3HNC5 E2BZN2 B4NX17 A0A1I8NP62 Q7JWF1 A0A034VR82 B4HMA2 A0A2B4SE12 A0A0L0BTW1 A0A1J1ITA4 T1P906 A0A0P6CVR2 A0A1D2MYL7 A0A0K8VCZ2 A0A3B0J4R6 A0A0J9R992 A0A1I8MF92 A0A091KYP4
A0A232FMI1 K7IXE6 A0A2J7PZJ7 J3JTE9 V5GUY2 A0A1B6C0L4 A0A182RUZ2 A0A151XJD1 A0A182NMH1 F4X3U3 A0A182Q739 A0A3L8DXW6 A0A026WNE4 A0A182M6W2 A0A182Y2X4 A0A158P050 A0A0P6J107 D2A0Q9 E2A3X9 A0A195E0P7 A0A195F1N3 A0A067RE05 A0A182PPA5 U5ES10 A0A182FE41 A0A195BFJ9 A0A084W2Q5 A0A2M4BHQ0 A0A1Y1KN64 W5JFW5 A0A182VKC9 A0A182IN15 A0A2M3Z244 A0A182XM95 A0A195CA52 A0A182WMD0 A0A182I230 A0A182GB59 A0A2A3E9F3 A0A2M4A6L1 A0A182TP92 Q7Q6E7 A0A2M4A6I3 A0A0B4J2M5 A0A182JRN1 A0A310SP62 A0A0A9WLX0 B0VZ76 R4WSM6 A0A1Q3FI44 T1DPN1 A0A0T6B555 A0A154NWP3 A0A182KPU7 A0A0L7QW90 J9JTH9 A0A0P4VSJ0 T1HF19 A0A2R5LJQ2 A0A0C9Q061 A0A1L8E0P0 A0A336KS65 A0A336M9M1 A0A224XDK6 A0A0V0G5A3 A0A2S2QCW4 Q171U3 E0VBI1 A0A1W4WSQ7 A0A2M4BIZ2 A0A1W4X2C3 A0A023FAJ2 W8BP19 A7RNA5 A0A1Z5KUI9 B3N6T6 A0A1S3HNC5 E2BZN2 B4NX17 A0A1I8NP62 Q7JWF1 A0A034VR82 B4HMA2 A0A2B4SE12 A0A0L0BTW1 A0A1J1ITA4 T1P906 A0A0P6CVR2 A0A1D2MYL7 A0A0K8VCZ2 A0A3B0J4R6 A0A0J9R992 A0A1I8MF92 A0A091KYP4
EC Number
1.8.1.4
Pubmed
19121390
26354079
28605547
22118469
28648823
20075255
+ More
22516182 23537049 21719571 30249741 24508170 25244985 21347285 26999592 18362917 19820115 20798317 24845553 24438588 28004739 20920257 23761445 26483478 12364791 14747013 17210077 25401762 26823975 23691247 20966253 27129103 17510324 20566863 25474469 24495485 17615350 28528879 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 26108605 27289101 22936249 25315136
22516182 23537049 21719571 30249741 24508170 25244985 21347285 26999592 18362917 19820115 20798317 24845553 24438588 28004739 20920257 23761445 26483478 12364791 14747013 17210077 25401762 26823975 23691247 20966253 27129103 17510324 20566863 25474469 24495485 17615350 28528879 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 26108605 27289101 22936249 25315136
EMBL
BABH01032924
BABH01032925
BABH01032926
KQ459337
KPJ01429.1
KY938822
+ More
ASA46463.1 RSAL01000059 RVE49687.1 AGBW02011894 OWR45962.1 GBYB01007221 JAG76988.1 NNAY01000020 OXU31882.1 AAZX01000010 NEVH01020335 PNF21740.1 BT126505 KB632306 AEE61469.1 ERL91896.1 GALX01002994 JAB65472.1 GEDC01030523 JAS06775.1 KQ982074 KYQ60522.1 GL888624 EGI58890.1 AXCN02000923 QOIP01000003 RLU25132.1 KK107144 EZA57555.1 AXCM01000805 ADTU01005358 GDUN01000272 JAN95647.1 KQ971338 EFA02551.1 GL436519 EFN71831.1 KQ979955 KYN18454.1 KQ981856 KYN34485.1 KK852566 KDR21248.1 GANO01002577 JAB57294.1 KQ976490 KYM83373.1 ATLV01019678 KE525277 KFB44499.1 GGFJ01003426 MBW52567.1 GEZM01081057 JAV61921.1 ADMH02001272 ETN63267.1 GGFM01001848 MBW22599.1 KQ978081 KYM97076.1 APCN01000109 JXUM01052081 KQ561718 KXJ77724.1 KZ288323 PBC28104.1 GGFK01002947 MBW36268.1 AAAB01008960 EAA11780.3 GGFK01003044 MBW36365.1 KQ760603 OAD59749.1 GBHO01035183 GBRD01003254 GDHC01008338 JAG08421.1 JAG62567.1 JAQ10291.1 DS231813 EDS25721.1 AK417712 BAN20927.1 GFDL01007831 JAV27214.1 GAMD01003204 JAA98386.1 LJIG01009737 KRT82482.1 KQ434775 KZC04106.1 KQ414716 KOC62872.1 ABLF02038315 ABLF02038324 ABLF02038335 ABLF02038338 GDKW01001680 JAI54915.1 ACPB03015449 GGLE01005529 MBY09655.1 GBYB01007223 JAG76990.1 GFDF01001784 JAV12300.1 UFQS01000777 UFQT01000777 SSX06811.1 SSX27156.1 UFQS01000761 UFQT01000761 SSX06698.1 SSX27044.1 GFTR01007352 JAW09074.1 GECL01002891 JAP03233.1 GGMS01006402 MBY75605.1 CH477446 EAT40764.1 EAT40765.1 DS235030 EEB10737.1 GGFJ01003812 MBW52953.1 GBBI01000240 JAC18472.1 GAMC01015106 JAB91449.1 DS469522 EDO47030.1 GFJQ02008275 JAV98694.1 CH954177 EDV58185.1 GL451645 EFN78847.1 CM000157 EDW89578.1 AE013599 AY113586 AAF58873.1 AAM29591.1 AFH07986.1 GAKP01014637 JAC44315.1 CH480816 EDW47179.1 LSMT01000120 PFX26675.1 JRES01001351 KNC23451.1 CVRI01000059 CRL03344.1 KA645179 AFP59808.1 GDIQ01087831 JAN06906.1 LJIJ01000379 ODM98143.1 GDHF01015889 GDHF01001522 JAI36425.1 JAI50792.1 OUUW01000001 SPP74553.1 CM002911 KMY92621.1 KK761821 KFP45177.1
ASA46463.1 RSAL01000059 RVE49687.1 AGBW02011894 OWR45962.1 GBYB01007221 JAG76988.1 NNAY01000020 OXU31882.1 AAZX01000010 NEVH01020335 PNF21740.1 BT126505 KB632306 AEE61469.1 ERL91896.1 GALX01002994 JAB65472.1 GEDC01030523 JAS06775.1 KQ982074 KYQ60522.1 GL888624 EGI58890.1 AXCN02000923 QOIP01000003 RLU25132.1 KK107144 EZA57555.1 AXCM01000805 ADTU01005358 GDUN01000272 JAN95647.1 KQ971338 EFA02551.1 GL436519 EFN71831.1 KQ979955 KYN18454.1 KQ981856 KYN34485.1 KK852566 KDR21248.1 GANO01002577 JAB57294.1 KQ976490 KYM83373.1 ATLV01019678 KE525277 KFB44499.1 GGFJ01003426 MBW52567.1 GEZM01081057 JAV61921.1 ADMH02001272 ETN63267.1 GGFM01001848 MBW22599.1 KQ978081 KYM97076.1 APCN01000109 JXUM01052081 KQ561718 KXJ77724.1 KZ288323 PBC28104.1 GGFK01002947 MBW36268.1 AAAB01008960 EAA11780.3 GGFK01003044 MBW36365.1 KQ760603 OAD59749.1 GBHO01035183 GBRD01003254 GDHC01008338 JAG08421.1 JAG62567.1 JAQ10291.1 DS231813 EDS25721.1 AK417712 BAN20927.1 GFDL01007831 JAV27214.1 GAMD01003204 JAA98386.1 LJIG01009737 KRT82482.1 KQ434775 KZC04106.1 KQ414716 KOC62872.1 ABLF02038315 ABLF02038324 ABLF02038335 ABLF02038338 GDKW01001680 JAI54915.1 ACPB03015449 GGLE01005529 MBY09655.1 GBYB01007223 JAG76990.1 GFDF01001784 JAV12300.1 UFQS01000777 UFQT01000777 SSX06811.1 SSX27156.1 UFQS01000761 UFQT01000761 SSX06698.1 SSX27044.1 GFTR01007352 JAW09074.1 GECL01002891 JAP03233.1 GGMS01006402 MBY75605.1 CH477446 EAT40764.1 EAT40765.1 DS235030 EEB10737.1 GGFJ01003812 MBW52953.1 GBBI01000240 JAC18472.1 GAMC01015106 JAB91449.1 DS469522 EDO47030.1 GFJQ02008275 JAV98694.1 CH954177 EDV58185.1 GL451645 EFN78847.1 CM000157 EDW89578.1 AE013599 AY113586 AAF58873.1 AAM29591.1 AFH07986.1 GAKP01014637 JAC44315.1 CH480816 EDW47179.1 LSMT01000120 PFX26675.1 JRES01001351 KNC23451.1 CVRI01000059 CRL03344.1 KA645179 AFP59808.1 GDIQ01087831 JAN06906.1 LJIJ01000379 ODM98143.1 GDHF01015889 GDHF01001522 JAI36425.1 JAI50792.1 OUUW01000001 SPP74553.1 CM002911 KMY92621.1 KK761821 KFP45177.1
Proteomes
UP000005204
UP000053268
UP000283053
UP000007151
UP000215335
UP000002358
+ More
UP000235965 UP000030742 UP000075900 UP000075809 UP000075884 UP000007755 UP000075886 UP000279307 UP000053097 UP000075883 UP000076408 UP000005205 UP000007266 UP000000311 UP000078492 UP000078541 UP000027135 UP000075885 UP000069272 UP000078540 UP000030765 UP000000673 UP000075903 UP000075880 UP000076407 UP000078542 UP000075920 UP000075840 UP000069940 UP000249989 UP000242457 UP000075902 UP000007062 UP000005203 UP000075881 UP000002320 UP000076502 UP000075882 UP000053825 UP000007819 UP000015103 UP000008820 UP000009046 UP000192223 UP000001593 UP000008711 UP000085678 UP000008237 UP000002282 UP000095300 UP000000803 UP000001292 UP000225706 UP000037069 UP000183832 UP000094527 UP000268350 UP000095301
UP000235965 UP000030742 UP000075900 UP000075809 UP000075884 UP000007755 UP000075886 UP000279307 UP000053097 UP000075883 UP000076408 UP000005205 UP000007266 UP000000311 UP000078492 UP000078541 UP000027135 UP000075885 UP000069272 UP000078540 UP000030765 UP000000673 UP000075903 UP000075880 UP000076407 UP000078542 UP000075920 UP000075840 UP000069940 UP000249989 UP000242457 UP000075902 UP000007062 UP000005203 UP000075881 UP000002320 UP000076502 UP000075882 UP000053825 UP000007819 UP000015103 UP000008820 UP000009046 UP000192223 UP000001593 UP000008711 UP000085678 UP000008237 UP000002282 UP000095300 UP000000803 UP000001292 UP000225706 UP000037069 UP000183832 UP000094527 UP000268350 UP000095301
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JIR9
A0A194Q7R1
A0A1Z2RRI8
A0A3S2PF61
A0A212EWT6
A0A0C9RK51
+ More
A0A232FMI1 K7IXE6 A0A2J7PZJ7 J3JTE9 V5GUY2 A0A1B6C0L4 A0A182RUZ2 A0A151XJD1 A0A182NMH1 F4X3U3 A0A182Q739 A0A3L8DXW6 A0A026WNE4 A0A182M6W2 A0A182Y2X4 A0A158P050 A0A0P6J107 D2A0Q9 E2A3X9 A0A195E0P7 A0A195F1N3 A0A067RE05 A0A182PPA5 U5ES10 A0A182FE41 A0A195BFJ9 A0A084W2Q5 A0A2M4BHQ0 A0A1Y1KN64 W5JFW5 A0A182VKC9 A0A182IN15 A0A2M3Z244 A0A182XM95 A0A195CA52 A0A182WMD0 A0A182I230 A0A182GB59 A0A2A3E9F3 A0A2M4A6L1 A0A182TP92 Q7Q6E7 A0A2M4A6I3 A0A0B4J2M5 A0A182JRN1 A0A310SP62 A0A0A9WLX0 B0VZ76 R4WSM6 A0A1Q3FI44 T1DPN1 A0A0T6B555 A0A154NWP3 A0A182KPU7 A0A0L7QW90 J9JTH9 A0A0P4VSJ0 T1HF19 A0A2R5LJQ2 A0A0C9Q061 A0A1L8E0P0 A0A336KS65 A0A336M9M1 A0A224XDK6 A0A0V0G5A3 A0A2S2QCW4 Q171U3 E0VBI1 A0A1W4WSQ7 A0A2M4BIZ2 A0A1W4X2C3 A0A023FAJ2 W8BP19 A7RNA5 A0A1Z5KUI9 B3N6T6 A0A1S3HNC5 E2BZN2 B4NX17 A0A1I8NP62 Q7JWF1 A0A034VR82 B4HMA2 A0A2B4SE12 A0A0L0BTW1 A0A1J1ITA4 T1P906 A0A0P6CVR2 A0A1D2MYL7 A0A0K8VCZ2 A0A3B0J4R6 A0A0J9R992 A0A1I8MF92 A0A091KYP4
A0A232FMI1 K7IXE6 A0A2J7PZJ7 J3JTE9 V5GUY2 A0A1B6C0L4 A0A182RUZ2 A0A151XJD1 A0A182NMH1 F4X3U3 A0A182Q739 A0A3L8DXW6 A0A026WNE4 A0A182M6W2 A0A182Y2X4 A0A158P050 A0A0P6J107 D2A0Q9 E2A3X9 A0A195E0P7 A0A195F1N3 A0A067RE05 A0A182PPA5 U5ES10 A0A182FE41 A0A195BFJ9 A0A084W2Q5 A0A2M4BHQ0 A0A1Y1KN64 W5JFW5 A0A182VKC9 A0A182IN15 A0A2M3Z244 A0A182XM95 A0A195CA52 A0A182WMD0 A0A182I230 A0A182GB59 A0A2A3E9F3 A0A2M4A6L1 A0A182TP92 Q7Q6E7 A0A2M4A6I3 A0A0B4J2M5 A0A182JRN1 A0A310SP62 A0A0A9WLX0 B0VZ76 R4WSM6 A0A1Q3FI44 T1DPN1 A0A0T6B555 A0A154NWP3 A0A182KPU7 A0A0L7QW90 J9JTH9 A0A0P4VSJ0 T1HF19 A0A2R5LJQ2 A0A0C9Q061 A0A1L8E0P0 A0A336KS65 A0A336M9M1 A0A224XDK6 A0A0V0G5A3 A0A2S2QCW4 Q171U3 E0VBI1 A0A1W4WSQ7 A0A2M4BIZ2 A0A1W4X2C3 A0A023FAJ2 W8BP19 A7RNA5 A0A1Z5KUI9 B3N6T6 A0A1S3HNC5 E2BZN2 B4NX17 A0A1I8NP62 Q7JWF1 A0A034VR82 B4HMA2 A0A2B4SE12 A0A0L0BTW1 A0A1J1ITA4 T1P906 A0A0P6CVR2 A0A1D2MYL7 A0A0K8VCZ2 A0A3B0J4R6 A0A0J9R992 A0A1I8MF92 A0A091KYP4
PDB
2GMJ
E-value=0,
Score=1926
Ontologies
GO
PANTHER
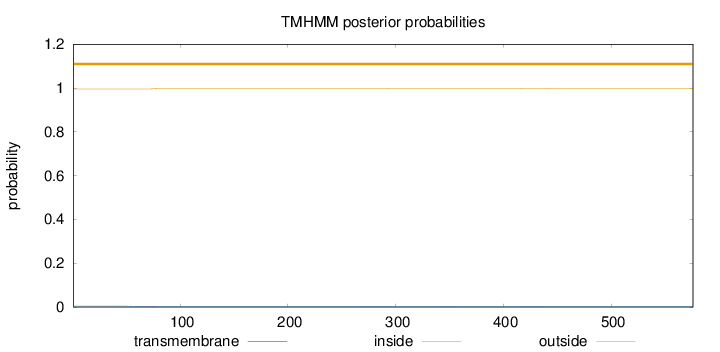
Topology
Length:
576
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0611100000000001
Exp number, first 60 AAs:
0.0067
Total prob of N-in:
0.00402
outside
1 - 576
Population Genetic Test Statistics
Pi
238.844172
Theta
177.302851
Tajima's D
1.770648
CLR
0.645873
CSRT
0.840857957102145
Interpretation
Uncertain
