Gene
KWMTBOMO08460 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009419
Annotation
PREDICTED:_malignant_T-cell-amplified_sequence_1_homolog_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 2.106 Mitochondrial Reliability : 1.529
Sequence
CDS
ATGCGTGTGTGTACGTTTGATGAAAAGGAAAGCATATCGGGCGTGCAACAGCTCAAGTCCTCGGTCCAGAAGGGCATAAGGGCTCGCCTTTTGGAACTGTACCCTCATTTGGACAACTACATTGACCAGATCTTACCCAAGAAGGATACGTTTAGGATTGTCAAATGTCACGATCACCTCGAGATAATGGTGAACAGTGCCGGCGACTTGCTATTCTTCAGGCATCGCGAGGGTCCCTGGATGCCAACCTTAAAACTACTGCACAAATATCCGTTCTTCGTTCCGATGCAGCAAGTTGACAAAGGCGCTATTCGCTTCGTGCTCAGCGGCGCGAACATCATGTGTCCGGGGCTGACTTCAGCCAACGCGAAGATGTCTCCGAGCGACAAGGGTCAAGTTGTCGCCATAATGGCCGAGGGGAAAGAACATGCTCTAGCTATCGGTATCACTACTCTCTCAACAGAAGACATAGCTAAAGTGAACAAAGGCGTCGGCGTTGAAAATTGTCATTATCTCAATGACGGGCTGTGGCAGATGAAGGCCGTGAAGTAA
Protein
MRVCTFDEKESISGVQQLKSSVQKGIRARLLELYPHLDNYIDQILPKKDTFRIVKCHDHLEIMVNSAGDLLFFRHREGPWMPTLKLLHKYPFFVPMQQVDKGAIRFVLSGANIMCPGLTSANAKMSPSDKGQVVAIMAEGKEHALAIGITTLSTEDIAKVNKGVGVENCHYLNDGLWQMKAVK
Summary
Uniprot
H9JIR8
A0A437BGW9
A0A1E1WKI6
S4PSZ5
A0A194Q8X8
A0A212EWW7
+ More
A0A0N1PJP5 A0A0J7NYM0 E2C440 A0A158N902 A0A151IBK9 A0A2A3ESB0 A0A088AGW6 E2A1G7 A0A151K0R0 F4WHK4 A0A154P6T4 A0A151J518 A0A151WNT6 A0A026VSQ6 A0A3L8DT69 A0A232F9X9 K7ISE9 A0A1B6LH62 B0WWD5 A0A1B6K1A1 A0A1Q3F8W8 A0A182NFV0 U5EN77 A0A0L7QWE8 A0A182H411 A0A067RD73 Q170Y4 A0A1B6D2P2 T1PE58 A0A0T6B832 A0A182XXB5 R4V0L8 A0A182M9G3 A0A182RQQ1 A0A1S3DK76 A0A1S4GID1 Q7Q9B2 D2A1L5 J3JY61 A0A182VXS7 A0A182QQW3 A0A1L8ECW0 A0A182JVP3 A0A1I8NQN0 A0A2J7QFS3 T1DNX1 A0A182XKD8 A0A182U207 A0A182HJK6 A0A0L0BZX1 A0A023F9L8 A0A182V4S8 A0A0A1XRX8 A0A0P4VTX8 R4FPP7 A0A182P8P1 A0A2M3Z9Y8 A0A2M4C0U4 W5JUH3 E0W295 A0A0K8TP68 A0A1Y1MUE5 A0A224XLM8 A0A0V0G6M8 A0A0M9A1H8 A0A2M3ZAE5 A0A2M4AK43 W8C2R0 A0A034UZ97 A0A2R7W179 A0A0K8U397 A0A1A9YJ55 A0A1A9UE35 A0A1B0BDG1 A0A1A9ZQF9 A0A182J2X6 A0A336MR50 A0A1B0DB80 A0A170ZKJ2 A0A2H8TZ10 C4WUE2 A0A2S2PKD5 A0A0A9YAF5 A0A1A9W791 A0A1L8DS23 A0A1B6I239 A0A1B0FLI1 B4M785 A0A2L2YB25 A0A2R5LI11 A0A1J1ILS9 B4JX87 B4NQ91 B4L1D5
A0A0N1PJP5 A0A0J7NYM0 E2C440 A0A158N902 A0A151IBK9 A0A2A3ESB0 A0A088AGW6 E2A1G7 A0A151K0R0 F4WHK4 A0A154P6T4 A0A151J518 A0A151WNT6 A0A026VSQ6 A0A3L8DT69 A0A232F9X9 K7ISE9 A0A1B6LH62 B0WWD5 A0A1B6K1A1 A0A1Q3F8W8 A0A182NFV0 U5EN77 A0A0L7QWE8 A0A182H411 A0A067RD73 Q170Y4 A0A1B6D2P2 T1PE58 A0A0T6B832 A0A182XXB5 R4V0L8 A0A182M9G3 A0A182RQQ1 A0A1S3DK76 A0A1S4GID1 Q7Q9B2 D2A1L5 J3JY61 A0A182VXS7 A0A182QQW3 A0A1L8ECW0 A0A182JVP3 A0A1I8NQN0 A0A2J7QFS3 T1DNX1 A0A182XKD8 A0A182U207 A0A182HJK6 A0A0L0BZX1 A0A023F9L8 A0A182V4S8 A0A0A1XRX8 A0A0P4VTX8 R4FPP7 A0A182P8P1 A0A2M3Z9Y8 A0A2M4C0U4 W5JUH3 E0W295 A0A0K8TP68 A0A1Y1MUE5 A0A224XLM8 A0A0V0G6M8 A0A0M9A1H8 A0A2M3ZAE5 A0A2M4AK43 W8C2R0 A0A034UZ97 A0A2R7W179 A0A0K8U397 A0A1A9YJ55 A0A1A9UE35 A0A1B0BDG1 A0A1A9ZQF9 A0A182J2X6 A0A336MR50 A0A1B0DB80 A0A170ZKJ2 A0A2H8TZ10 C4WUE2 A0A2S2PKD5 A0A0A9YAF5 A0A1A9W791 A0A1L8DS23 A0A1B6I239 A0A1B0FLI1 B4M785 A0A2L2YB25 A0A2R5LI11 A0A1J1ILS9 B4JX87 B4NQ91 B4L1D5
Pubmed
19121390
23622113
26354079
22118469
20798317
21347285
+ More
21719571 24508170 30249741 28648823 20075255 26483478 24845553 17510324 25315136 25244985 12364791 18362917 19820115 22516182 26108605 25474469 25830018 27129103 20920257 23761445 20566863 26369729 28004739 24495485 25348373 25401762 26823975 17994087 26561354
21719571 24508170 30249741 28648823 20075255 26483478 24845553 17510324 25315136 25244985 12364791 18362917 19820115 22516182 26108605 25474469 25830018 27129103 20920257 23761445 20566863 26369729 28004739 24495485 25348373 25401762 26823975 17994087 26561354
EMBL
BABH01032922
BABH01032923
BABH01032924
RSAL01000059
RVE49686.1
GDQN01003524
+ More
JAT87530.1 GAIX01012928 JAA79632.1 KQ459337 KPJ01430.1 AGBW02011894 OWR45961.1 KQ459986 KPJ18816.1 LBMM01000785 KMQ97500.1 GL452364 EFN77426.1 ADTU01009011 KQ978095 KYM97025.1 KZ288189 PBC34645.1 GL435766 EFN72640.1 KQ981229 KYN44343.1 GL888161 EGI66358.1 KQ434827 KZC07587.1 KQ980063 KYN17955.1 KQ982907 KYQ49451.1 KK108326 EZA46757.1 QOIP01000004 RLU23363.1 NNAY01000561 OXU27656.1 GEBQ01016924 JAT23053.1 DS232143 DS232270 EDS36027.1 EDS38821.1 GECU01017841 GECU01002495 JAS89865.1 JAT05212.1 GFDL01011076 JAV23969.1 GANO01000706 JAB59165.1 KQ414713 KOC62953.1 JXUM01108588 KQ565245 KXJ71190.1 KK852536 KDR21831.1 CH477463 EAT40525.1 GEDC01017415 JAS19883.1 KA646238 AFP60867.1 LJIG01009221 KRT83514.1 KC632348 AGM32162.1 AXCM01010076 AAAB01008904 EAA09663.4 KQ971338 EFA02688.1 BT128185 AEE63146.1 AXCN02000249 GFDG01002323 JAV16476.1 NEVH01014850 PNF27440.1 GAMD01002480 JAA99110.1 APCN01002178 JRES01001179 KNC24769.1 GBBI01000627 JAC18085.1 GBXI01000560 JAD13732.1 GDKW01000605 JAI55990.1 ACPB03004193 GAHY01000769 JAA76741.1 GGFM01004592 MBW25343.1 GGFJ01009805 MBW58946.1 ADMH02000413 ETN66615.1 DS235875 EEB19751.1 GDAI01001662 JAI15941.1 GEZM01020354 JAV89292.1 GFTR01003030 JAW13396.1 GECL01002515 JAP03609.1 KQ435794 KOX73879.1 GGFM01004766 MBW25517.1 GGFK01007834 MBW41155.1 GAMC01000333 JAC06223.1 GAKP01023359 JAC35599.1 KK854138 PTY11945.1 GDHF01031529 JAI20785.1 JXJN01012467 UFQT01001633 SSX31263.1 AJVK01004663 GEMB01002085 JAS01094.1 GFXV01007709 MBW19514.1 ABLF02041081 AK341028 BAH71512.1 GGMR01016707 MBY29326.1 GBHO01014460 GBRD01009733 GDHC01015249 JAG29144.1 JAG56091.1 JAQ03380.1 GFDF01004883 JAV09201.1 GECU01026717 JAS80989.1 CCAG010005843 CH940653 EDW62652.1 IAAA01009873 LAA04560.1 GGLE01004831 MBY08957.1 CVRI01000050 CRK99417.1 CH916376 EDV95363.1 CH964291 EDW86316.1 CH933810 EDW06656.1
JAT87530.1 GAIX01012928 JAA79632.1 KQ459337 KPJ01430.1 AGBW02011894 OWR45961.1 KQ459986 KPJ18816.1 LBMM01000785 KMQ97500.1 GL452364 EFN77426.1 ADTU01009011 KQ978095 KYM97025.1 KZ288189 PBC34645.1 GL435766 EFN72640.1 KQ981229 KYN44343.1 GL888161 EGI66358.1 KQ434827 KZC07587.1 KQ980063 KYN17955.1 KQ982907 KYQ49451.1 KK108326 EZA46757.1 QOIP01000004 RLU23363.1 NNAY01000561 OXU27656.1 GEBQ01016924 JAT23053.1 DS232143 DS232270 EDS36027.1 EDS38821.1 GECU01017841 GECU01002495 JAS89865.1 JAT05212.1 GFDL01011076 JAV23969.1 GANO01000706 JAB59165.1 KQ414713 KOC62953.1 JXUM01108588 KQ565245 KXJ71190.1 KK852536 KDR21831.1 CH477463 EAT40525.1 GEDC01017415 JAS19883.1 KA646238 AFP60867.1 LJIG01009221 KRT83514.1 KC632348 AGM32162.1 AXCM01010076 AAAB01008904 EAA09663.4 KQ971338 EFA02688.1 BT128185 AEE63146.1 AXCN02000249 GFDG01002323 JAV16476.1 NEVH01014850 PNF27440.1 GAMD01002480 JAA99110.1 APCN01002178 JRES01001179 KNC24769.1 GBBI01000627 JAC18085.1 GBXI01000560 JAD13732.1 GDKW01000605 JAI55990.1 ACPB03004193 GAHY01000769 JAA76741.1 GGFM01004592 MBW25343.1 GGFJ01009805 MBW58946.1 ADMH02000413 ETN66615.1 DS235875 EEB19751.1 GDAI01001662 JAI15941.1 GEZM01020354 JAV89292.1 GFTR01003030 JAW13396.1 GECL01002515 JAP03609.1 KQ435794 KOX73879.1 GGFM01004766 MBW25517.1 GGFK01007834 MBW41155.1 GAMC01000333 JAC06223.1 GAKP01023359 JAC35599.1 KK854138 PTY11945.1 GDHF01031529 JAI20785.1 JXJN01012467 UFQT01001633 SSX31263.1 AJVK01004663 GEMB01002085 JAS01094.1 GFXV01007709 MBW19514.1 ABLF02041081 AK341028 BAH71512.1 GGMR01016707 MBY29326.1 GBHO01014460 GBRD01009733 GDHC01015249 JAG29144.1 JAG56091.1 JAQ03380.1 GFDF01004883 JAV09201.1 GECU01026717 JAS80989.1 CCAG010005843 CH940653 EDW62652.1 IAAA01009873 LAA04560.1 GGLE01004831 MBY08957.1 CVRI01000050 CRK99417.1 CH916376 EDV95363.1 CH964291 EDW86316.1 CH933810 EDW06656.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000007151
UP000053240
UP000036403
+ More
UP000008237 UP000005205 UP000078542 UP000242457 UP000005203 UP000000311 UP000078541 UP000007755 UP000076502 UP000078492 UP000075809 UP000053097 UP000279307 UP000215335 UP000002358 UP000002320 UP000075884 UP000053825 UP000069940 UP000249989 UP000027135 UP000008820 UP000095301 UP000076408 UP000075883 UP000075900 UP000079169 UP000007062 UP000007266 UP000075920 UP000075886 UP000075881 UP000095300 UP000235965 UP000076407 UP000075902 UP000075840 UP000037069 UP000075903 UP000015103 UP000075885 UP000000673 UP000009046 UP000053105 UP000092443 UP000078200 UP000092460 UP000092445 UP000075880 UP000092462 UP000007819 UP000091820 UP000092444 UP000008792 UP000183832 UP000001070 UP000007798 UP000009192
UP000008237 UP000005205 UP000078542 UP000242457 UP000005203 UP000000311 UP000078541 UP000007755 UP000076502 UP000078492 UP000075809 UP000053097 UP000279307 UP000215335 UP000002358 UP000002320 UP000075884 UP000053825 UP000069940 UP000249989 UP000027135 UP000008820 UP000095301 UP000076408 UP000075883 UP000075900 UP000079169 UP000007062 UP000007266 UP000075920 UP000075886 UP000075881 UP000095300 UP000235965 UP000076407 UP000075902 UP000075840 UP000037069 UP000075903 UP000015103 UP000075885 UP000000673 UP000009046 UP000053105 UP000092443 UP000078200 UP000092460 UP000092445 UP000075880 UP000092462 UP000007819 UP000091820 UP000092444 UP000008792 UP000183832 UP000001070 UP000007798 UP000009192
PRIDE
Interpro
SUPFAM
SSF88697
SSF88697
Gene 3D
ProteinModelPortal
H9JIR8
A0A437BGW9
A0A1E1WKI6
S4PSZ5
A0A194Q8X8
A0A212EWW7
+ More
A0A0N1PJP5 A0A0J7NYM0 E2C440 A0A158N902 A0A151IBK9 A0A2A3ESB0 A0A088AGW6 E2A1G7 A0A151K0R0 F4WHK4 A0A154P6T4 A0A151J518 A0A151WNT6 A0A026VSQ6 A0A3L8DT69 A0A232F9X9 K7ISE9 A0A1B6LH62 B0WWD5 A0A1B6K1A1 A0A1Q3F8W8 A0A182NFV0 U5EN77 A0A0L7QWE8 A0A182H411 A0A067RD73 Q170Y4 A0A1B6D2P2 T1PE58 A0A0T6B832 A0A182XXB5 R4V0L8 A0A182M9G3 A0A182RQQ1 A0A1S3DK76 A0A1S4GID1 Q7Q9B2 D2A1L5 J3JY61 A0A182VXS7 A0A182QQW3 A0A1L8ECW0 A0A182JVP3 A0A1I8NQN0 A0A2J7QFS3 T1DNX1 A0A182XKD8 A0A182U207 A0A182HJK6 A0A0L0BZX1 A0A023F9L8 A0A182V4S8 A0A0A1XRX8 A0A0P4VTX8 R4FPP7 A0A182P8P1 A0A2M3Z9Y8 A0A2M4C0U4 W5JUH3 E0W295 A0A0K8TP68 A0A1Y1MUE5 A0A224XLM8 A0A0V0G6M8 A0A0M9A1H8 A0A2M3ZAE5 A0A2M4AK43 W8C2R0 A0A034UZ97 A0A2R7W179 A0A0K8U397 A0A1A9YJ55 A0A1A9UE35 A0A1B0BDG1 A0A1A9ZQF9 A0A182J2X6 A0A336MR50 A0A1B0DB80 A0A170ZKJ2 A0A2H8TZ10 C4WUE2 A0A2S2PKD5 A0A0A9YAF5 A0A1A9W791 A0A1L8DS23 A0A1B6I239 A0A1B0FLI1 B4M785 A0A2L2YB25 A0A2R5LI11 A0A1J1ILS9 B4JX87 B4NQ91 B4L1D5
A0A0N1PJP5 A0A0J7NYM0 E2C440 A0A158N902 A0A151IBK9 A0A2A3ESB0 A0A088AGW6 E2A1G7 A0A151K0R0 F4WHK4 A0A154P6T4 A0A151J518 A0A151WNT6 A0A026VSQ6 A0A3L8DT69 A0A232F9X9 K7ISE9 A0A1B6LH62 B0WWD5 A0A1B6K1A1 A0A1Q3F8W8 A0A182NFV0 U5EN77 A0A0L7QWE8 A0A182H411 A0A067RD73 Q170Y4 A0A1B6D2P2 T1PE58 A0A0T6B832 A0A182XXB5 R4V0L8 A0A182M9G3 A0A182RQQ1 A0A1S3DK76 A0A1S4GID1 Q7Q9B2 D2A1L5 J3JY61 A0A182VXS7 A0A182QQW3 A0A1L8ECW0 A0A182JVP3 A0A1I8NQN0 A0A2J7QFS3 T1DNX1 A0A182XKD8 A0A182U207 A0A182HJK6 A0A0L0BZX1 A0A023F9L8 A0A182V4S8 A0A0A1XRX8 A0A0P4VTX8 R4FPP7 A0A182P8P1 A0A2M3Z9Y8 A0A2M4C0U4 W5JUH3 E0W295 A0A0K8TP68 A0A1Y1MUE5 A0A224XLM8 A0A0V0G6M8 A0A0M9A1H8 A0A2M3ZAE5 A0A2M4AK43 W8C2R0 A0A034UZ97 A0A2R7W179 A0A0K8U397 A0A1A9YJ55 A0A1A9UE35 A0A1B0BDG1 A0A1A9ZQF9 A0A182J2X6 A0A336MR50 A0A1B0DB80 A0A170ZKJ2 A0A2H8TZ10 C4WUE2 A0A2S2PKD5 A0A0A9YAF5 A0A1A9W791 A0A1L8DS23 A0A1B6I239 A0A1B0FLI1 B4M785 A0A2L2YB25 A0A2R5LI11 A0A1J1ILS9 B4JX87 B4NQ91 B4L1D5
PDB
6MS4
E-value=1.97858e-65,
Score=628
Ontologies
GO
PANTHER
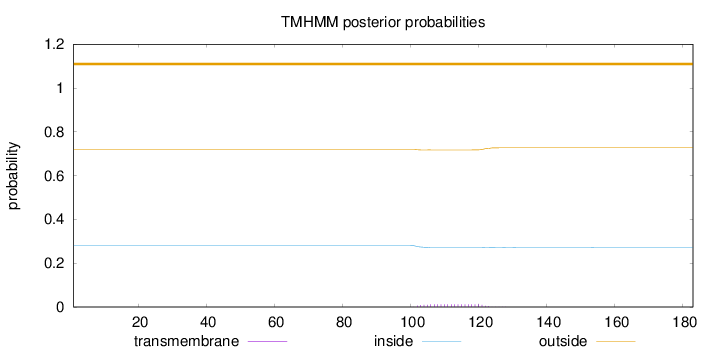
Topology
Subcellular location
Cytoplasm
Length:
183
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.24344
Exp number, first 60 AAs:
0.0001
Total prob of N-in:
0.28047
outside
1 - 183
Population Genetic Test Statistics
Pi
256.925442
Theta
174.883741
Tajima's D
1.489396
CLR
0.386728
CSRT
0.786060696965152
Interpretation
Uncertain
