Gene
KWMTBOMO08459
Pre Gene Modal
BGIBMGA009418
Annotation
PREDICTED:_adiponectin_receptor_protein_[Papilio_polytes]
Full name
Adiponectin receptor protein
Location in the cell
PlasmaMembrane Reliability : 4.528
Sequence
CDS
ATGTGGGAAGTGGAATCTGATATTGGATCCCAAAGCGTGTCCTCCGATGGATTGCGACGCAGGCAAGGATGGGATCCCGATGCGGAGAGCCTTGCCTCTCAAATGGACGAGCTCGACGAAGTCCTCGCAGAAGAAGAAGAAGGATGTCCGCTGCCTTCAACACCGGAGGATCAACATCTGTTGGATGCCGAAATGGCTGAAGTTTTGAAAGCTGGCGTGTTATCCGATGAGATTGATTTGGGAGCACTGGCCCATAACGCCGCCGAACAGGCCGAGGAATTCGTTCGCAAAGTATGGGAGGCGTCGTGGAACGTGTGCCATTTCAGACATTTGCCACGTTGGCTACAAGACAACGATTATTTACACAAAGGACATAGACCGCCTCTACCTTCGTTCAGCGCATGTTTTGCATCAATTTTCCGAATCCACACTGAGACAGGTAACATTTGGACGCATCTCCTTGGCTGCGTGGCATTCATTGGCGTCGCGATCTACTTTCTGTCTCGTCCATCTATCGAAATTCAAATGCAAGAGAAAGTTATATTCGGTGTTTTTTTTGTCGGCGCTATCGTATGCCTCGGTTTTTCTTTTGCCTATCACACACTGTACTGCCACTCCGAGATGGTCGGAAAGCTGTTCTCAAAGCTGGATTATTGTGGAATAGCATTGCTCATCATGGGCTCCTTTGTTCCATGGTTGTACTACAGTTTCTACTGCCACTACAGACCGAAGATCATATACCTATCTGTAGTAGTTGTTTTAGGAATTTTGTCAATAATAGTGTCTTTGTGGGATAGATTCTCAGAACCTCGACTAAGACCTCTCAGAGCAGGAGTTTTTATGGGCTTTGGTTTGTCTGGTATAGTCCCAGCAATTCACTATGGGATTACCGAAGGCTGGTTCAGTCAAGTCAGCAAAGCATCATTGGGCTGGTTAGTTTTGATGGGATTGCTCTATATCTTAGGTGCCATGTTCTATGCCTTAAGAGTGCCAGAACGTTGGTTCCCTGGCAAATGTGATATTTGGTTTCAGTCCCATCAAATATTCCATGTTCTTGTGATTGTAGCTGCTTTTGTACATTACCACGGTATAAGCGAATTGGCATCTTACAGAGTCACAGTAGGAGAGTGCTCCATGCCACCAACATCAATGGCATTTTAG
Protein
MWEVESDIGSQSVSSDGLRRRQGWDPDAESLASQMDELDEVLAEEEEGCPLPSTPEDQHLLDAEMAEVLKAGVLSDEIDLGALAHNAAEQAEEFVRKVWEASWNVCHFRHLPRWLQDNDYLHKGHRPPLPSFSACFASIFRIHTETGNIWTHLLGCVAFIGVAIYFLSRPSIEIQMQEKVIFGVFFVGAIVCLGFSFAYHTLYCHSEMVGKLFSKLDYCGIALLIMGSFVPWLYYSFYCHYRPKIIYLSVVVVLGILSIIVSLWDRFSEPRLRPLRAGVFMGFGLSGIVPAIHYGITEGWFSQVSKASLGWLVLMGLLYILGAMFYALRVPERWFPGKCDIWFQSHQIFHVLVIVAAFVHYHGISELASYRVTVGECSMPPTSMAF
Summary
Description
Adiponectin receptor. In insulin-producing cells, regulates insulin secretion and controls glucose and lipid metabolism.
Similarity
Belongs to the ADIPOR family.
Keywords
Alternative splicing
Cell membrane
Complete proteome
Fatty acid metabolism
Lipid metabolism
Membrane
Metal-binding
Receptor
Reference proteome
Transmembrane
Transmembrane helix
Zinc
Feature
chain Adiponectin receptor protein
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9JIR7
A0A1E1W0N3
A0A2A4JND7
A0A2H1W7A5
A0A0L7LBW4
A0A0N1IGF4
+ More
A0A194QD99 A0A212EWT3 S4PGR1 D8VKL0 D8VKQ2 D8VKM0 D8VKK0 D8VKP0 D8VKP1 A0SY07 U5EVN7 A0A182MB76 A0A182RZK7 A0A182QSN1 A0A182XIJ7 A0A182HYN1 A0A336MAM0 A0A182KZK5 A0A2J7QMW4 A0A182VPC6 A0A182JXL0 A0A182PP86 A0A182TN93 Q7PQ98 A0A182JGW8 A0A182W3F1 B0W6M8 A0A084WA12 A0A182Y8T3 A0A034W669 A0A0K8VY50 T1DE99 A0A1L8DK54 A0A1Q3FFP5 A0A2M3YY90 A0A0A1X2L3 A0A182FSS7 A0A2M4BNQ8 W5J494 A0A023ET23 T1DRF4 A0A2M4BNT6 W8C5A6 A0A2M4A0H1 A0A182GIH3 Q16FL3 A0A067RD16 B4K9B8 A0A1B0CBV8 B4M0I2 A0A1L8EEA7 A0A1I8PCZ2 A0A0K8TR76 A0A0M4F554 B4JV41 A0A1I8MMI4 A0A1I8MMI7 A0A1I8MMJ3 A0A0L0BZY7 A0A023ERV3 A4V392 Q9VCY8 B4PMB4 D3TLT6 A0A3B0K567 A0A1W4W3X4 A0A182GJW9 B3LZZ9 I5AN13 B4NJS6 A0A2A3EFL6 A0A088ASM1 A0A026WBW4 F4WAE1 A0A195FSM4 A0A158NKX0 E2C3U2 A0A195E484 A0A151WMI4 A0A0M9AAN5 E2B0K9 A0A0L7QQS9 B4HEP5 A0A195CSF5 B3P8N5 A0A232F1C5 B4R0M1 A0A0Q9X0Y8 A0A0Q9WSU0 A0A1L8EED1 A0A1I8PD00 A0A0Q9WS27 A0A1L8EEB0 A0A0Q9WGU3
A0A194QD99 A0A212EWT3 S4PGR1 D8VKL0 D8VKQ2 D8VKM0 D8VKK0 D8VKP0 D8VKP1 A0SY07 U5EVN7 A0A182MB76 A0A182RZK7 A0A182QSN1 A0A182XIJ7 A0A182HYN1 A0A336MAM0 A0A182KZK5 A0A2J7QMW4 A0A182VPC6 A0A182JXL0 A0A182PP86 A0A182TN93 Q7PQ98 A0A182JGW8 A0A182W3F1 B0W6M8 A0A084WA12 A0A182Y8T3 A0A034W669 A0A0K8VY50 T1DE99 A0A1L8DK54 A0A1Q3FFP5 A0A2M3YY90 A0A0A1X2L3 A0A182FSS7 A0A2M4BNQ8 W5J494 A0A023ET23 T1DRF4 A0A2M4BNT6 W8C5A6 A0A2M4A0H1 A0A182GIH3 Q16FL3 A0A067RD16 B4K9B8 A0A1B0CBV8 B4M0I2 A0A1L8EEA7 A0A1I8PCZ2 A0A0K8TR76 A0A0M4F554 B4JV41 A0A1I8MMI4 A0A1I8MMI7 A0A1I8MMJ3 A0A0L0BZY7 A0A023ERV3 A4V392 Q9VCY8 B4PMB4 D3TLT6 A0A3B0K567 A0A1W4W3X4 A0A182GJW9 B3LZZ9 I5AN13 B4NJS6 A0A2A3EFL6 A0A088ASM1 A0A026WBW4 F4WAE1 A0A195FSM4 A0A158NKX0 E2C3U2 A0A195E484 A0A151WMI4 A0A0M9AAN5 E2B0K9 A0A0L7QQS9 B4HEP5 A0A195CSF5 B3P8N5 A0A232F1C5 B4R0M1 A0A0Q9X0Y8 A0A0Q9WSU0 A0A1L8EED1 A0A1I8PD00 A0A0Q9WS27 A0A1L8EEB0 A0A0Q9WGU3
Pubmed
19121390
26227816
26354079
22118469
23622113
20442862
+ More
20966253 12364791 14747013 17210077 24438588 25244985 25348373 24330624 25830018 20920257 23761445 24945155 24495485 26483478 17510324 24845553 17994087 18057021 26369729 25315136 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12537569 23874700 17550304 20353571 15632085 24508170 30249741 21719571 21347285 20798317 28648823
20966253 12364791 14747013 17210077 24438588 25244985 25348373 24330624 25830018 20920257 23761445 24945155 24495485 26483478 17510324 24845553 17994087 18057021 26369729 25315136 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12537569 23874700 17550304 20353571 15632085 24508170 30249741 21719571 21347285 20798317 28648823
EMBL
BABH01032921
GDQN01010531
JAT80523.1
NWSH01000906
PCG73585.1
ODYU01006786
+ More
SOQ48960.1 JTDY01001864 KOB72691.1 KQ459986 KPJ18817.1 KQ459337 KPJ01431.1 AGBW02011894 OWR45960.1 GAIX01006100 JAA86460.1 GQ987617 GQ987618 GQ987619 GQ987620 GQ987621 GQ987622 GQ987623 GQ987624 GQ987625 GQ987626 ADI81284.1 GQ987659 GQ987660 GQ987661 GQ987662 GQ987663 GQ987664 GQ987665 GQ987666 ADI81326.1 GQ987627 GQ987628 GQ987629 GQ987630 GQ987631 GQ987632 GQ987633 GQ987634 GQ987635 GQ987636 GQ987637 GQ987638 GQ987639 GQ987640 GQ987641 GQ987642 GQ987643 GQ987644 GQ987645 GQ987646 ADI81294.1 GQ987607 GQ987608 GQ987609 GQ987610 GQ987611 GQ987612 GQ987613 GQ987614 GQ987615 GQ987616 ADI81274.1 GQ987647 GQ987649 GQ987650 GQ987651 GQ987652 GQ987653 GQ987654 GQ987655 GQ987656 GQ987657 GQ987658 ADI81314.1 GQ987648 ADI81315.1 EF062308 ABK57116.2 GANO01003326 JAB56545.1 AXCM01001884 AXCN02001612 APCN01005391 UFQT01000309 SSX23088.1 NEVH01013202 PNF29926.1 AAAB01008898 EAA09141.6 DS231848 EDS36702.1 ATLV01021997 KE525327 KFB47056.1 GAKP01009160 GAKP01009159 JAC49792.1 GDHF01008555 JAI43759.1 GALA01001122 JAA93730.1 GFDF01007268 JAV06816.1 GFDL01008675 JAV26370.1 GGFM01000488 MBW21239.1 GBXI01009166 GBXI01000472 JAD05126.1 JAD13820.1 GGFJ01005493 MBW54634.1 ADMH02002125 ETN58701.1 GAPW01001647 JAC11951.1 GAMD01000650 JAB00941.1 GGFJ01005523 MBW54664.1 GAMC01004994 GAMC01004993 GAMC01004992 JAC01562.1 GGFK01000924 MBW34245.1 JXUM01066108 KQ562381 KXJ76032.1 CH478416 EAT33030.1 KK852715 KDR17851.1 CH933806 EDW15550.1 KRG01614.1 AJWK01005874 AJWK01005875 CH940650 EDW68361.1 KRF83802.1 KRF83803.1 GFDG01001768 JAV17031.1 GDAI01000726 JAI16877.1 CP012526 ALC46607.1 CH916374 EDV91361.1 JRES01001095 KNC25615.1 GAPW01001646 JAC11952.1 AE014297 BT133262 BT133272 AAN13909.1 AFC35446.1 AFC35457.1 AFH06557.1 AGB96224.1 BI585801 BT001487 CM000160 EDW98019.2 KRK04068.1 KRK04069.1 EZ422388 ADD18664.1 OUUW01000005 SPP80766.1 JXUM01013038 JXUM01013039 JXUM01013040 JXUM01013041 JXUM01013042 JXUM01013043 KQ560367 KXJ82779.1 CH902617 EDV44189.2 KPU80675.1 CM000070 EIM52348.1 CH964272 EDW85038.1 KZ288271 PBC29781.1 KK107283 QOIP01000001 EZA53550.1 RLU27495.1 RLU27496.1 GL888048 EGI68824.1 KQ981281 KYN43297.1 ADTU01018979 GL452364 EFN77328.1 KQ979701 KYN19664.1 KQ982937 KYQ49102.1 KQ435710 KOX79926.1 GL444634 EFN60769.1 KQ414785 KOC60982.1 CH480815 EDW43206.1 KQ977306 KYN03631.1 CH954182 EDV54130.1 NNAY01001281 OXU24505.1 CM000364 EDX13944.1 KRG01613.1 KRF83806.1 GFDG01001727 JAV17072.1 KRF83807.1 GFDG01001752 JAV17047.1 KRF83805.1
SOQ48960.1 JTDY01001864 KOB72691.1 KQ459986 KPJ18817.1 KQ459337 KPJ01431.1 AGBW02011894 OWR45960.1 GAIX01006100 JAA86460.1 GQ987617 GQ987618 GQ987619 GQ987620 GQ987621 GQ987622 GQ987623 GQ987624 GQ987625 GQ987626 ADI81284.1 GQ987659 GQ987660 GQ987661 GQ987662 GQ987663 GQ987664 GQ987665 GQ987666 ADI81326.1 GQ987627 GQ987628 GQ987629 GQ987630 GQ987631 GQ987632 GQ987633 GQ987634 GQ987635 GQ987636 GQ987637 GQ987638 GQ987639 GQ987640 GQ987641 GQ987642 GQ987643 GQ987644 GQ987645 GQ987646 ADI81294.1 GQ987607 GQ987608 GQ987609 GQ987610 GQ987611 GQ987612 GQ987613 GQ987614 GQ987615 GQ987616 ADI81274.1 GQ987647 GQ987649 GQ987650 GQ987651 GQ987652 GQ987653 GQ987654 GQ987655 GQ987656 GQ987657 GQ987658 ADI81314.1 GQ987648 ADI81315.1 EF062308 ABK57116.2 GANO01003326 JAB56545.1 AXCM01001884 AXCN02001612 APCN01005391 UFQT01000309 SSX23088.1 NEVH01013202 PNF29926.1 AAAB01008898 EAA09141.6 DS231848 EDS36702.1 ATLV01021997 KE525327 KFB47056.1 GAKP01009160 GAKP01009159 JAC49792.1 GDHF01008555 JAI43759.1 GALA01001122 JAA93730.1 GFDF01007268 JAV06816.1 GFDL01008675 JAV26370.1 GGFM01000488 MBW21239.1 GBXI01009166 GBXI01000472 JAD05126.1 JAD13820.1 GGFJ01005493 MBW54634.1 ADMH02002125 ETN58701.1 GAPW01001647 JAC11951.1 GAMD01000650 JAB00941.1 GGFJ01005523 MBW54664.1 GAMC01004994 GAMC01004993 GAMC01004992 JAC01562.1 GGFK01000924 MBW34245.1 JXUM01066108 KQ562381 KXJ76032.1 CH478416 EAT33030.1 KK852715 KDR17851.1 CH933806 EDW15550.1 KRG01614.1 AJWK01005874 AJWK01005875 CH940650 EDW68361.1 KRF83802.1 KRF83803.1 GFDG01001768 JAV17031.1 GDAI01000726 JAI16877.1 CP012526 ALC46607.1 CH916374 EDV91361.1 JRES01001095 KNC25615.1 GAPW01001646 JAC11952.1 AE014297 BT133262 BT133272 AAN13909.1 AFC35446.1 AFC35457.1 AFH06557.1 AGB96224.1 BI585801 BT001487 CM000160 EDW98019.2 KRK04068.1 KRK04069.1 EZ422388 ADD18664.1 OUUW01000005 SPP80766.1 JXUM01013038 JXUM01013039 JXUM01013040 JXUM01013041 JXUM01013042 JXUM01013043 KQ560367 KXJ82779.1 CH902617 EDV44189.2 KPU80675.1 CM000070 EIM52348.1 CH964272 EDW85038.1 KZ288271 PBC29781.1 KK107283 QOIP01000001 EZA53550.1 RLU27495.1 RLU27496.1 GL888048 EGI68824.1 KQ981281 KYN43297.1 ADTU01018979 GL452364 EFN77328.1 KQ979701 KYN19664.1 KQ982937 KYQ49102.1 KQ435710 KOX79926.1 GL444634 EFN60769.1 KQ414785 KOC60982.1 CH480815 EDW43206.1 KQ977306 KYN03631.1 CH954182 EDV54130.1 NNAY01001281 OXU24505.1 CM000364 EDX13944.1 KRG01613.1 KRF83806.1 GFDG01001727 JAV17072.1 KRF83807.1 GFDG01001752 JAV17047.1 KRF83805.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053240
UP000053268
UP000007151
+ More
UP000075883 UP000075900 UP000075886 UP000076407 UP000075840 UP000075882 UP000235965 UP000075903 UP000075881 UP000075885 UP000075902 UP000007062 UP000075880 UP000075920 UP000002320 UP000030765 UP000076408 UP000069272 UP000000673 UP000069940 UP000249989 UP000008820 UP000027135 UP000009192 UP000092461 UP000008792 UP000095300 UP000092553 UP000001070 UP000095301 UP000037069 UP000000803 UP000002282 UP000268350 UP000192221 UP000007801 UP000001819 UP000007798 UP000242457 UP000005203 UP000053097 UP000279307 UP000007755 UP000078541 UP000005205 UP000008237 UP000078492 UP000075809 UP000053105 UP000000311 UP000053825 UP000001292 UP000078542 UP000008711 UP000215335 UP000000304
UP000075883 UP000075900 UP000075886 UP000076407 UP000075840 UP000075882 UP000235965 UP000075903 UP000075881 UP000075885 UP000075902 UP000007062 UP000075880 UP000075920 UP000002320 UP000030765 UP000076408 UP000069272 UP000000673 UP000069940 UP000249989 UP000008820 UP000027135 UP000009192 UP000092461 UP000008792 UP000095300 UP000092553 UP000001070 UP000095301 UP000037069 UP000000803 UP000002282 UP000268350 UP000192221 UP000007801 UP000001819 UP000007798 UP000242457 UP000005203 UP000053097 UP000279307 UP000007755 UP000078541 UP000005205 UP000008237 UP000078492 UP000075809 UP000053105 UP000000311 UP000053825 UP000001292 UP000078542 UP000008711 UP000215335 UP000000304
ProteinModelPortal
H9JIR7
A0A1E1W0N3
A0A2A4JND7
A0A2H1W7A5
A0A0L7LBW4
A0A0N1IGF4
+ More
A0A194QD99 A0A212EWT3 S4PGR1 D8VKL0 D8VKQ2 D8VKM0 D8VKK0 D8VKP0 D8VKP1 A0SY07 U5EVN7 A0A182MB76 A0A182RZK7 A0A182QSN1 A0A182XIJ7 A0A182HYN1 A0A336MAM0 A0A182KZK5 A0A2J7QMW4 A0A182VPC6 A0A182JXL0 A0A182PP86 A0A182TN93 Q7PQ98 A0A182JGW8 A0A182W3F1 B0W6M8 A0A084WA12 A0A182Y8T3 A0A034W669 A0A0K8VY50 T1DE99 A0A1L8DK54 A0A1Q3FFP5 A0A2M3YY90 A0A0A1X2L3 A0A182FSS7 A0A2M4BNQ8 W5J494 A0A023ET23 T1DRF4 A0A2M4BNT6 W8C5A6 A0A2M4A0H1 A0A182GIH3 Q16FL3 A0A067RD16 B4K9B8 A0A1B0CBV8 B4M0I2 A0A1L8EEA7 A0A1I8PCZ2 A0A0K8TR76 A0A0M4F554 B4JV41 A0A1I8MMI4 A0A1I8MMI7 A0A1I8MMJ3 A0A0L0BZY7 A0A023ERV3 A4V392 Q9VCY8 B4PMB4 D3TLT6 A0A3B0K567 A0A1W4W3X4 A0A182GJW9 B3LZZ9 I5AN13 B4NJS6 A0A2A3EFL6 A0A088ASM1 A0A026WBW4 F4WAE1 A0A195FSM4 A0A158NKX0 E2C3U2 A0A195E484 A0A151WMI4 A0A0M9AAN5 E2B0K9 A0A0L7QQS9 B4HEP5 A0A195CSF5 B3P8N5 A0A232F1C5 B4R0M1 A0A0Q9X0Y8 A0A0Q9WSU0 A0A1L8EED1 A0A1I8PD00 A0A0Q9WS27 A0A1L8EEB0 A0A0Q9WGU3
A0A194QD99 A0A212EWT3 S4PGR1 D8VKL0 D8VKQ2 D8VKM0 D8VKK0 D8VKP0 D8VKP1 A0SY07 U5EVN7 A0A182MB76 A0A182RZK7 A0A182QSN1 A0A182XIJ7 A0A182HYN1 A0A336MAM0 A0A182KZK5 A0A2J7QMW4 A0A182VPC6 A0A182JXL0 A0A182PP86 A0A182TN93 Q7PQ98 A0A182JGW8 A0A182W3F1 B0W6M8 A0A084WA12 A0A182Y8T3 A0A034W669 A0A0K8VY50 T1DE99 A0A1L8DK54 A0A1Q3FFP5 A0A2M3YY90 A0A0A1X2L3 A0A182FSS7 A0A2M4BNQ8 W5J494 A0A023ET23 T1DRF4 A0A2M4BNT6 W8C5A6 A0A2M4A0H1 A0A182GIH3 Q16FL3 A0A067RD16 B4K9B8 A0A1B0CBV8 B4M0I2 A0A1L8EEA7 A0A1I8PCZ2 A0A0K8TR76 A0A0M4F554 B4JV41 A0A1I8MMI4 A0A1I8MMI7 A0A1I8MMJ3 A0A0L0BZY7 A0A023ERV3 A4V392 Q9VCY8 B4PMB4 D3TLT6 A0A3B0K567 A0A1W4W3X4 A0A182GJW9 B3LZZ9 I5AN13 B4NJS6 A0A2A3EFL6 A0A088ASM1 A0A026WBW4 F4WAE1 A0A195FSM4 A0A158NKX0 E2C3U2 A0A195E484 A0A151WMI4 A0A0M9AAN5 E2B0K9 A0A0L7QQS9 B4HEP5 A0A195CSF5 B3P8N5 A0A232F1C5 B4R0M1 A0A0Q9X0Y8 A0A0Q9WSU0 A0A1L8EED1 A0A1I8PD00 A0A0Q9WS27 A0A1L8EEB0 A0A0Q9WGU3
PDB
5LXA
E-value=6.88747e-89,
Score=834
Ontologies
GO
PANTHER
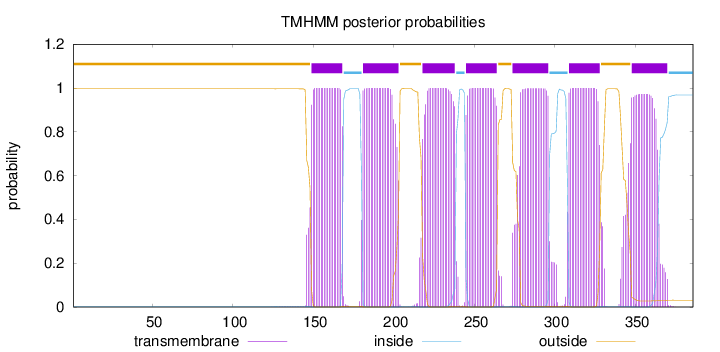
Topology
Subcellular location
Cell membrane
Length:
386
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
147.79407
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00357
outside
1 - 148
TMhelix
149 - 168
inside
169 - 180
TMhelix
181 - 203
outside
204 - 217
TMhelix
218 - 238
inside
239 - 244
TMhelix
245 - 264
outside
265 - 273
TMhelix
274 - 296
inside
297 - 308
TMhelix
309 - 328
outside
329 - 347
TMhelix
348 - 370
inside
371 - 386
Population Genetic Test Statistics
Pi
342.000053
Theta
245.014401
Tajima's D
1.231977
CLR
0
CSRT
0.72426378681066
Interpretation
Uncertain
