Pre Gene Modal
BGIBMGA009395
Annotation
PREDICTED:_WD_repeat-containing_protein_18_[Plutella_xylostella]
Location in the cell
Cytoplasmic Reliability : 1.446 PlasmaMembrane Reliability : 1.735
Sequence
CDS
ATGGCAAATTTACTTGAATTACTGGTTACCGCTGACTGCAACAATCCTTTGTGGACAGCAAGCATCTGGGATTCACATATCGGTACGAATCTCATGACGTACAAAGGTGGAGGTACTGCCGAATCAAAAACGCTTTCTTTCATCGGTAACGATTATATAGCAGCAGTAGAAAAGACTAAACCAATATTACACATATGGCCCTTGAATTCACATCAGCCGGTCCAAGGAATGCGGTTTATTTTACCCGGAAAAGCGAGTGCCTTGTCTATAACCCGAGACGGCTCCTACATATGTGCTGGAATCGAAGAGAAAATCTACTTGTGGCAGACTGCGTCGGGAAATCTGTTGACTATAATCAGCCGTCACTATCAGAAAGTTGTATCATTAAAATTCACACCAGATGGAAAGTATTTCATATCAGCGGCTGAAGATGGAATGGTAATGGTTTGGTCGCTTGCATCTGTGGCAGCACATCCAGAAATCGATCTTGTGACGCAGTCGTCTGCTGGCCAACATGACCCAGTTCATATCTTCTCTGATCATTCTCTACCTGTAACTGATTTGTTTATAAGTAAATTTGGTATGCATGGACGTCTGTGTACTGTGTCTAGTGATAGGACTTGTAAAATATATGATCTATCTACTGGAGAGATGTTATTGAATCTAGTCTTTGATGTTCCTCTGTCGGCTGTCACTATGGATGTTTTAGAATTGAATGTGTTTGTTGGAAGCAATGAAGGTAAAATATATCAGTTCAGCCTTGCCCATCCACCTAGGAGCAGAGATTTATTGATCAATTCTGAAAACGGATCTCCGGTCTTCACGTTACATACTAAAGCTATAACATGTCTCTCTGTCTCATTAGATGGTGAAACTCTCATGTCAGGTTCCAATGATGAGCATGTTATACTATGGCACATACCTAGTCGACAGCCAATTAGAACTATAAGACACAAAGGGCCAATTACAAATGGATTTTTTACTGTAAACCTTCCAGCTATTTTTCAATCTGAATTTTGTCCTGACATTGTTCTACATAGTTTGGAGAGGACACTTGAAAAAGATTCTGATGAAGCTACTGAAATAGAAGTCTTGATAAATAAGAAAATTAGTTTTTGGCCAGAACAAAATTCTTATGAAATCAAAGAAATAATACAACAAACTGAGACTTCATTTATGGGACAAGAAAAGAAATGGAAAAATGAAATAGAAACTTTAAAGGCAGTAAATGCAAAACTTTATGCATTTTCTATAAATAAAGCAATAGATGCAGTTCCATTAGTGAATGAATCAATTATAAAAAAGGCTAAAAAGAAGAAAAACAATAAGAAAAATAAGTAA
Protein
MANLLELLVTADCNNPLWTASIWDSHIGTNLMTYKGGGTAESKTLSFIGNDYIAAVEKTKPILHIWPLNSHQPVQGMRFILPGKASALSITRDGSYICAGIEEKIYLWQTASGNLLTIISRHYQKVVSLKFTPDGKYFISAAEDGMVMVWSLASVAAHPEIDLVTQSSAGQHDPVHIFSDHSLPVTDLFISKFGMHGRLCTVSSDRTCKIYDLSTGEMLLNLVFDVPLSAVTMDVLELNVFVGSNEGKIYQFSLAHPPRSRDLLINSENGSPVFTLHTKAITCLSVSLDGETLMSGSNDEHVILWHIPSRQPIRTIRHKGPITNGFFTVNLPAIFQSEFCPDIVLHSLERTLEKDSDEATEIEVLINKKISFWPEQNSYEIKEIIQQTETSFMGQEKKWKNEIETLKAVNAKLYAFSINKAIDAVPLVNESIIKKAKKKKNNKKNK
Summary
Uniprot
H9JIP4
A0A2A4JPA1
A0A2H1W7A6
A0A0L7LB43
S4PEZ2
A0A194Q7B3
+ More
A0A0N1PHC8 A0A212EWU2 A0A067RMJ8 A0A2J7PNM0 A0A2J7RSA7 A0A182G866 B0WSI0 Q16HG2 A0A1S4G142 A0A1Q3F1G0 A0A1J1HNN8 E0V9A7 A0A0L0CJ24 A0A087ZUL9 A0A2A3ET14 A0A154P879 A0A0A1X0R4 A0A1A9VEK2 A0A1I8M9B2 W8C4L3 A0A232FAG9 A0A0M9A2A2 A0A3Q0J3P3 K7ISF3 A0A034VFH4 D3TS49 A0A1B0FEZ0 F4X3R1 A0A1I8P2U0 A0A0K8WE85 A0A1B0APM6 A0A158P035 A0A195E037 A0A0L7R356 A0A1S3IZC0 C3YQL5 A0A0J7L526 A0A2H8TZS8 A0A195F353 E9IT01 A0A2M4AMP6 A0A1L8DC06 A0A2M4CSA1 A0A1B0CSY7 A0A1L8DCA2 A0A2M4CSQ3 A0A1A8FNN5 A0A1B6C927 A0A0C9QZS8 A0A2M4BNX7 A0A2M4BNU6 A0A1A8PRY2 A0A1A8QK52 A0A1A8IJV3 E2AYH7 A0A195CQP2 A0A3P9D1B0 A0A3P8QJ79 A0A2M4BNT1 A0A151XJW0 J9K534 A0A2U9C1B0 I3JA47 A0A1A8B3S2 A0A1A8CP30 A0A3B4G3I6 A0A3Q2V6E5 A0A026WP58 A0A2S2PES9 B3MXH7 A0A3B5ADH4 B3P9N3 A0A310SG52 A0A2U9C2W2 B4Q1L2 A0A3Q1CUJ0 A0A2I4DD27 A0A3Q3MC66 A0A0N7ZPI6 A0A3P8S136 A0A164Q6L8 B4I9J9 A0A3Q3RB18 W5UJ28 Q9W550 B4N279 A0A1A7YTQ1 F1LR39 A0A182YC96 Q8SX33 A0A1W4ZKL7 A0A3B4G3K8
A0A0N1PHC8 A0A212EWU2 A0A067RMJ8 A0A2J7PNM0 A0A2J7RSA7 A0A182G866 B0WSI0 Q16HG2 A0A1S4G142 A0A1Q3F1G0 A0A1J1HNN8 E0V9A7 A0A0L0CJ24 A0A087ZUL9 A0A2A3ET14 A0A154P879 A0A0A1X0R4 A0A1A9VEK2 A0A1I8M9B2 W8C4L3 A0A232FAG9 A0A0M9A2A2 A0A3Q0J3P3 K7ISF3 A0A034VFH4 D3TS49 A0A1B0FEZ0 F4X3R1 A0A1I8P2U0 A0A0K8WE85 A0A1B0APM6 A0A158P035 A0A195E037 A0A0L7R356 A0A1S3IZC0 C3YQL5 A0A0J7L526 A0A2H8TZS8 A0A195F353 E9IT01 A0A2M4AMP6 A0A1L8DC06 A0A2M4CSA1 A0A1B0CSY7 A0A1L8DCA2 A0A2M4CSQ3 A0A1A8FNN5 A0A1B6C927 A0A0C9QZS8 A0A2M4BNX7 A0A2M4BNU6 A0A1A8PRY2 A0A1A8QK52 A0A1A8IJV3 E2AYH7 A0A195CQP2 A0A3P9D1B0 A0A3P8QJ79 A0A2M4BNT1 A0A151XJW0 J9K534 A0A2U9C1B0 I3JA47 A0A1A8B3S2 A0A1A8CP30 A0A3B4G3I6 A0A3Q2V6E5 A0A026WP58 A0A2S2PES9 B3MXH7 A0A3B5ADH4 B3P9N3 A0A310SG52 A0A2U9C2W2 B4Q1L2 A0A3Q1CUJ0 A0A2I4DD27 A0A3Q3MC66 A0A0N7ZPI6 A0A3P8S136 A0A164Q6L8 B4I9J9 A0A3Q3RB18 W5UJ28 Q9W550 B4N279 A0A1A7YTQ1 F1LR39 A0A182YC96 Q8SX33 A0A1W4ZKL7 A0A3B4G3K8
Pubmed
19121390
26227816
23622113
26354079
22118469
24845553
+ More
26483478 17510324 20566863 26108605 25830018 25315136 24495485 28648823 20075255 25348373 20353571 21719571 21347285 18563158 21282665 20798317 25186727 24508170 30249741 17994087 17550304 23127152 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15057822 15632090 25244985
26483478 17510324 20566863 26108605 25830018 25315136 24495485 28648823 20075255 25348373 20353571 21719571 21347285 18563158 21282665 20798317 25186727 24508170 30249741 17994087 17550304 23127152 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15057822 15632090 25244985
EMBL
BABH01032921
NWSH01000906
PCG73586.1
ODYU01006786
SOQ48961.1
JTDY01001864
+ More
KOB72693.1 GAIX01004277 JAA88283.1 KQ459337 KPJ01432.1 KQ459986 KPJ18818.1 AGBW02011894 OWR45959.1 KK852536 KDR21830.1 NEVH01023953 PNF17916.1 NEVH01000263 PNF43726.1 JXUM01047489 KQ561521 KXJ78295.1 DS232072 EDS33910.1 CH478175 EAT33685.1 GFDL01013717 JAV21328.1 CVRI01000014 CRK89661.1 DS234991 EEB09963.1 JRES01000333 KNC32227.1 KZ288185 PBC34868.1 KQ434839 KZC08053.1 GBXI01009962 JAD04330.1 GAMC01009526 JAB97029.1 NNAY01000561 OXU27655.1 KQ435758 KOX75825.1 GAKP01018407 JAC40545.1 EZ424251 ADD20527.1 CCAG010003465 GL888624 EGI58858.1 GDHF01003149 JAI49165.1 JXJN01001491 ADTU01005289 KQ979955 KYN18483.1 KQ414663 KOC65307.1 GG666543 EEN57371.1 LBMM01000566 KMQ98052.1 GFXV01007456 MBW19261.1 KQ981856 KYN34514.1 GL765434 EFZ16264.1 GGFK01008733 MBW42054.1 GFDF01010086 JAV03998.1 GGFL01004032 MBW68210.1 AJWK01026782 GFDF01010087 JAV03997.1 GGFL01004033 MBW68211.1 HAEB01013222 HAEC01003627 SBQ59749.1 GEDC01027364 GEDC01022028 GEDC01002003 JAS09934.1 JAS15270.1 JAS35295.1 GBYB01001220 JAG70987.1 GGFJ01005532 MBW54673.1 GGFJ01005533 MBW54674.1 HAEF01015180 HAEG01009153 SBR83749.1 HAEH01012007 HAEI01016153 SBR93703.1 HAED01011170 HAEE01015330 SBQ97468.1 GL443910 EFN61512.1 KQ977394 KYN03063.1 GGFJ01005531 MBW54672.1 KQ982074 KYQ60490.1 ABLF02036171 CP026254 AWP10405.1 AERX01026330 AERX01026331 AERX01026332 AERX01026333 HADY01022861 HAEJ01001599 SBP61346.1 HADZ01017591 SBP81532.1 KK107139 QOIP01000003 EZA57842.1 RLU24786.1 GGMR01015331 MBY27950.1 CH902630 EDV38442.1 CH954183 EDV45529.1 KQ768090 OAD53219.1 AWP10403.1 CM000162 EDX01453.1 GDIP01225249 JAI98152.1 LRGB01002451 KZS07477.1 CH480825 EDW43880.1 JT416284 AHH41996.1 AE014298 BT133493 AAF45681.1 AFI71923.1 CH963925 EDW78468.1 HADW01008945 HADX01011339 SBP33571.1 AABR07055844 CH474029 EDL89362.1 AY094880 AAM11233.1
KOB72693.1 GAIX01004277 JAA88283.1 KQ459337 KPJ01432.1 KQ459986 KPJ18818.1 AGBW02011894 OWR45959.1 KK852536 KDR21830.1 NEVH01023953 PNF17916.1 NEVH01000263 PNF43726.1 JXUM01047489 KQ561521 KXJ78295.1 DS232072 EDS33910.1 CH478175 EAT33685.1 GFDL01013717 JAV21328.1 CVRI01000014 CRK89661.1 DS234991 EEB09963.1 JRES01000333 KNC32227.1 KZ288185 PBC34868.1 KQ434839 KZC08053.1 GBXI01009962 JAD04330.1 GAMC01009526 JAB97029.1 NNAY01000561 OXU27655.1 KQ435758 KOX75825.1 GAKP01018407 JAC40545.1 EZ424251 ADD20527.1 CCAG010003465 GL888624 EGI58858.1 GDHF01003149 JAI49165.1 JXJN01001491 ADTU01005289 KQ979955 KYN18483.1 KQ414663 KOC65307.1 GG666543 EEN57371.1 LBMM01000566 KMQ98052.1 GFXV01007456 MBW19261.1 KQ981856 KYN34514.1 GL765434 EFZ16264.1 GGFK01008733 MBW42054.1 GFDF01010086 JAV03998.1 GGFL01004032 MBW68210.1 AJWK01026782 GFDF01010087 JAV03997.1 GGFL01004033 MBW68211.1 HAEB01013222 HAEC01003627 SBQ59749.1 GEDC01027364 GEDC01022028 GEDC01002003 JAS09934.1 JAS15270.1 JAS35295.1 GBYB01001220 JAG70987.1 GGFJ01005532 MBW54673.1 GGFJ01005533 MBW54674.1 HAEF01015180 HAEG01009153 SBR83749.1 HAEH01012007 HAEI01016153 SBR93703.1 HAED01011170 HAEE01015330 SBQ97468.1 GL443910 EFN61512.1 KQ977394 KYN03063.1 GGFJ01005531 MBW54672.1 KQ982074 KYQ60490.1 ABLF02036171 CP026254 AWP10405.1 AERX01026330 AERX01026331 AERX01026332 AERX01026333 HADY01022861 HAEJ01001599 SBP61346.1 HADZ01017591 SBP81532.1 KK107139 QOIP01000003 EZA57842.1 RLU24786.1 GGMR01015331 MBY27950.1 CH902630 EDV38442.1 CH954183 EDV45529.1 KQ768090 OAD53219.1 AWP10403.1 CM000162 EDX01453.1 GDIP01225249 JAI98152.1 LRGB01002451 KZS07477.1 CH480825 EDW43880.1 JT416284 AHH41996.1 AE014298 BT133493 AAF45681.1 AFI71923.1 CH963925 EDW78468.1 HADW01008945 HADX01011339 SBP33571.1 AABR07055844 CH474029 EDL89362.1 AY094880 AAM11233.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053268
UP000053240
UP000007151
+ More
UP000027135 UP000235965 UP000069940 UP000249989 UP000002320 UP000008820 UP000183832 UP000009046 UP000037069 UP000005203 UP000242457 UP000076502 UP000078200 UP000095301 UP000215335 UP000053105 UP000079169 UP000002358 UP000092444 UP000007755 UP000095300 UP000092460 UP000005205 UP000078492 UP000053825 UP000085678 UP000001554 UP000036403 UP000078541 UP000092461 UP000000311 UP000078542 UP000265160 UP000265100 UP000075809 UP000007819 UP000246464 UP000005207 UP000261460 UP000264840 UP000053097 UP000279307 UP000007801 UP000261400 UP000008711 UP000002282 UP000257160 UP000192220 UP000261640 UP000265080 UP000076858 UP000001292 UP000261600 UP000000803 UP000007798 UP000002494 UP000076408 UP000192224
UP000027135 UP000235965 UP000069940 UP000249989 UP000002320 UP000008820 UP000183832 UP000009046 UP000037069 UP000005203 UP000242457 UP000076502 UP000078200 UP000095301 UP000215335 UP000053105 UP000079169 UP000002358 UP000092444 UP000007755 UP000095300 UP000092460 UP000005205 UP000078492 UP000053825 UP000085678 UP000001554 UP000036403 UP000078541 UP000092461 UP000000311 UP000078542 UP000265160 UP000265100 UP000075809 UP000007819 UP000246464 UP000005207 UP000261460 UP000264840 UP000053097 UP000279307 UP000007801 UP000261400 UP000008711 UP000002282 UP000257160 UP000192220 UP000261640 UP000265080 UP000076858 UP000001292 UP000261600 UP000000803 UP000007798 UP000002494 UP000076408 UP000192224
Interpro
IPR011047
Quinoprotein_ADH-like_supfam
+ More
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR036322 WD40_repeat_dom_sf
IPR035007 ANKLE2
IPR036770 Ankyrin_rpt-contain_sf
IPR020683 Ankyrin_rpt-contain_dom
IPR002110 Ankyrin_rpt
IPR019775 WD40_repeat_CS
IPR020472 G-protein_beta_WD-40_rep
IPR026987 Wdr18_C_dom
IPR024977 Apc4_WD40_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR036322 WD40_repeat_dom_sf
IPR035007 ANKLE2
IPR036770 Ankyrin_rpt-contain_sf
IPR020683 Ankyrin_rpt-contain_dom
IPR002110 Ankyrin_rpt
IPR019775 WD40_repeat_CS
IPR020472 G-protein_beta_WD-40_rep
IPR026987 Wdr18_C_dom
IPR024977 Apc4_WD40_dom
Gene 3D
CDD
ProteinModelPortal
H9JIP4
A0A2A4JPA1
A0A2H1W7A6
A0A0L7LB43
S4PEZ2
A0A194Q7B3
+ More
A0A0N1PHC8 A0A212EWU2 A0A067RMJ8 A0A2J7PNM0 A0A2J7RSA7 A0A182G866 B0WSI0 Q16HG2 A0A1S4G142 A0A1Q3F1G0 A0A1J1HNN8 E0V9A7 A0A0L0CJ24 A0A087ZUL9 A0A2A3ET14 A0A154P879 A0A0A1X0R4 A0A1A9VEK2 A0A1I8M9B2 W8C4L3 A0A232FAG9 A0A0M9A2A2 A0A3Q0J3P3 K7ISF3 A0A034VFH4 D3TS49 A0A1B0FEZ0 F4X3R1 A0A1I8P2U0 A0A0K8WE85 A0A1B0APM6 A0A158P035 A0A195E037 A0A0L7R356 A0A1S3IZC0 C3YQL5 A0A0J7L526 A0A2H8TZS8 A0A195F353 E9IT01 A0A2M4AMP6 A0A1L8DC06 A0A2M4CSA1 A0A1B0CSY7 A0A1L8DCA2 A0A2M4CSQ3 A0A1A8FNN5 A0A1B6C927 A0A0C9QZS8 A0A2M4BNX7 A0A2M4BNU6 A0A1A8PRY2 A0A1A8QK52 A0A1A8IJV3 E2AYH7 A0A195CQP2 A0A3P9D1B0 A0A3P8QJ79 A0A2M4BNT1 A0A151XJW0 J9K534 A0A2U9C1B0 I3JA47 A0A1A8B3S2 A0A1A8CP30 A0A3B4G3I6 A0A3Q2V6E5 A0A026WP58 A0A2S2PES9 B3MXH7 A0A3B5ADH4 B3P9N3 A0A310SG52 A0A2U9C2W2 B4Q1L2 A0A3Q1CUJ0 A0A2I4DD27 A0A3Q3MC66 A0A0N7ZPI6 A0A3P8S136 A0A164Q6L8 B4I9J9 A0A3Q3RB18 W5UJ28 Q9W550 B4N279 A0A1A7YTQ1 F1LR39 A0A182YC96 Q8SX33 A0A1W4ZKL7 A0A3B4G3K8
A0A0N1PHC8 A0A212EWU2 A0A067RMJ8 A0A2J7PNM0 A0A2J7RSA7 A0A182G866 B0WSI0 Q16HG2 A0A1S4G142 A0A1Q3F1G0 A0A1J1HNN8 E0V9A7 A0A0L0CJ24 A0A087ZUL9 A0A2A3ET14 A0A154P879 A0A0A1X0R4 A0A1A9VEK2 A0A1I8M9B2 W8C4L3 A0A232FAG9 A0A0M9A2A2 A0A3Q0J3P3 K7ISF3 A0A034VFH4 D3TS49 A0A1B0FEZ0 F4X3R1 A0A1I8P2U0 A0A0K8WE85 A0A1B0APM6 A0A158P035 A0A195E037 A0A0L7R356 A0A1S3IZC0 C3YQL5 A0A0J7L526 A0A2H8TZS8 A0A195F353 E9IT01 A0A2M4AMP6 A0A1L8DC06 A0A2M4CSA1 A0A1B0CSY7 A0A1L8DCA2 A0A2M4CSQ3 A0A1A8FNN5 A0A1B6C927 A0A0C9QZS8 A0A2M4BNX7 A0A2M4BNU6 A0A1A8PRY2 A0A1A8QK52 A0A1A8IJV3 E2AYH7 A0A195CQP2 A0A3P9D1B0 A0A3P8QJ79 A0A2M4BNT1 A0A151XJW0 J9K534 A0A2U9C1B0 I3JA47 A0A1A8B3S2 A0A1A8CP30 A0A3B4G3I6 A0A3Q2V6E5 A0A026WP58 A0A2S2PES9 B3MXH7 A0A3B5ADH4 B3P9N3 A0A310SG52 A0A2U9C2W2 B4Q1L2 A0A3Q1CUJ0 A0A2I4DD27 A0A3Q3MC66 A0A0N7ZPI6 A0A3P8S136 A0A164Q6L8 B4I9J9 A0A3Q3RB18 W5UJ28 Q9W550 B4N279 A0A1A7YTQ1 F1LR39 A0A182YC96 Q8SX33 A0A1W4ZKL7 A0A3B4G3K8
PDB
5AFU
E-value=2.3694e-10,
Score=158
Ontologies
GO
PANTHER
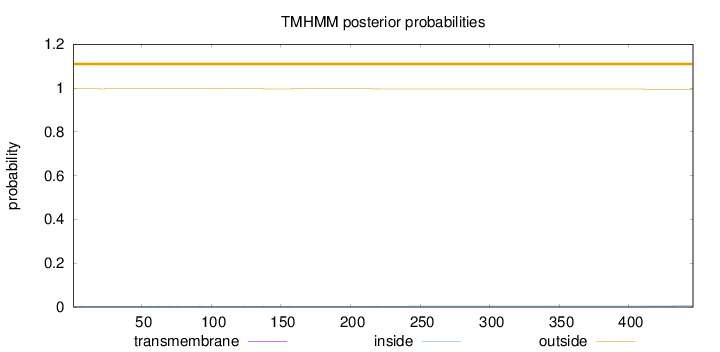
Topology
Length:
446
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05491
Exp number, first 60 AAs:
0.00106
Total prob of N-in:
0.00380
outside
1 - 446
Population Genetic Test Statistics
Pi
259.912861
Theta
231.53385
Tajima's D
0.479335
CLR
0.100176
CSRT
0.504074796260187
Interpretation
Uncertain
