Gene
KWMTBOMO08451
Pre Gene Modal
BGIBMGA009398
Annotation
PREDICTED:_histone_deacetylase_11_[Papilio_xuthus]
Full name
Histone deacetylase 11
Location in the cell
Cytoplasmic Reliability : 2.333
Sequence
CDS
ATGACAAGTCTGTACTTCGATATAAGCCAAGAGCAATGGCCAATAGTATATGATGACAAATACAATGTTTCGGTTTTTGGTCTCGAGAAGTTTCATGTATTTGACGCTAAAAAGTGGCGGAATATTGTTCAGTATTTAAGGAATGCACAACTTATAACGGATGAACGCCTAGTGAAGCCAAAAGAAGCCCAGAAGAGTGATTTGCTTGTTGTCCATACAAAGCGGTATCTCAAATCTTTGAATTGGAGCGCAAGAGTGGCTATAATCGCAGAAGTACCACTTGTATCTTTATTACCAAATTTCCTGGTTCAATACGCTTATTTAAAGCCGATGAGGTTACAAACAGGTGGTTCAGTTCTTTGTGGTAAGCTGGCATTGGAGCGTGGGTGGGCTATCAATGTGGGTGGTGGCTTCCACCACTGCAGTGCTCAAAAAGGCGAAGGTTTCTGTCCGTATGCTGACATAACGCTGCTGATACGGTTCCTCATGATGAATAACTTAATAGAGACTGCTATGATTGTCGATTTGGATGCTCATCAGGGAAATGGTTACGAAAAGGATTTCCTGGGTGTTCCCGAAGTATACATAATGGATATGTACAATAGAAATATTTTCCCGAAAGACAAAGAAGCGAAGAAGGCGATACGAAGGAAAGTGGAACTCGGGAATCTGGTTGAGGACATGGAATATATGCTGAAATTGCGATTGAATTTAAAAGCAGCCTTTAATGAATTCAAACCGGATATAGTTGTGTACAACGCGGGCACTGATATTTTGGACACAGATCCTTTGGGACACATGAGCATAAGTGAATGTGGGATAATAAAACGGGACGAATACGTATTCGAGATGTGTAAAGGTCTCCGCGTGCCCATAGTCATGCTGACCAGCGGCGGCTATCTGAGGAGGACCGCGCGGATCATAGCCGAGTCGATCATTAACTTGCACGGCAAAGGTCTAATATACGGAAGCTGTAAAGACAGTAAATGGTAA
Protein
MTSLYFDISQEQWPIVYDDKYNVSVFGLEKFHVFDAKKWRNIVQYLRNAQLITDERLVKPKEAQKSDLLVVHTKRYLKSLNWSARVAIIAEVPLVSLLPNFLVQYAYLKPMRLQTGGSVLCGKLALERGWAINVGGGFHHCSAQKGEGFCPYADITLLIRFLMMNNLIETAMIVDLDAHQGNGYEKDFLGVPEVYIMDMYNRNIFPKDKEAKKAIRRKVELGNLVEDMEYMLKLRLNLKAAFNEFKPDIVVYNAGTDILDTDPLGHMSISECGIIKRDEYVFEMCKGLRVPIVMLTSGGYLRRTARIIAESIINLHGKGLIYGSCKDSKW
Summary
Description
Responsible for the deacetylation of lysine residues on the N-terminal part of the core histones (H2A, H2B, H3 and H4). Histone deacetylation gives a tag for epigenetic repression and plays an important role in transcriptional regulation, cell cycle progression and developmental events. Histone deacetylases act via the formation of large multiprotein complexes.
Responsible for the deacetylation of lysine residues on the N-terminal part of the core histones (H2A, H2B, H3 and H4). Histone deacetylation gives a tag for epigenetic repression and plays an important role in transcriptional regulation, cell cycle progression and developmental events. Histone deacetylases act via the formation of large multiprotein complexes (By similarity).
Responsible for the deacetylation of lysine residues on the N-terminal part of the core histones (H2A, H2B, H3 and H4). Histone deacetylation gives a tag for epigenetic repression and plays an important role in transcriptional regulation, cell cycle progression and developmental events. Histone deacetylases act via the formation of large multiprotein complexes (By similarity).
Catalytic Activity
Hydrolysis of an N(6)-acetyl-lysine residue of a histone to yield a deacetylated histone.
Subunit
Interacts with HDAC6.
Miscellaneous
Its activity is inhibited by trapoxin, a known histone deacetylase inhibitor.
Similarity
Belongs to the histone deacetylase family.
Keywords
Alternative splicing
Chromatin regulator
Complete proteome
Hydrolase
Nucleus
Reference proteome
Repressor
Transcription
Transcription regulation
Feature
chain Histone deacetylase 11
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9JIP7
A0A3S2TLX4
A0A2H1VNL7
A0A194Q6P7
A0A0N1PHA1
A0A1B6F569
+ More
A0A067R947 A0A2J7PGD8 A0A2P8XLX9 I3LTU6 A0A1B6MAN5 A0A2Y9ENA6 A0A1Y1LWP4 A0A1B6DMB6 A0A384A6S9 A0A341CY01 G3SLU4 A0A2Y9MM77 A0A099ZPM0 A0A093I875 A0A2K6U715 A0A091EX69 A0A3Q7Q521 A0A2Y9HGJ3 A0A2K6U773 A0A2Y9D5Z2 K7DAK7 A0A024R2I1 Q96DB2 A0A2K5SAC0 G3QXL9 A0A3Q2KRU9 F6WJS2 A0A2J8NNX6 A0A2J8TVT1 A0A2K5JRW1 A0A2K6QLH9 A0A2K6KMM0 F7HYS2 A0A3Q2I1P4 A0A2K5ETR4 A0A452QXR8 A0A2K5JRT8 A0A3B3WIE7 A0A091TKR1 A0A093CY31 A0A093BLT0 A0A2J8TVT9 A0A3Q7V7T1 A0A2U3VLA8 H2P9B0 A0A3Q7RNE1 A0A091Q2D1 A0A402EFM4 M3W7T9 A0A2I2UGE1 A0A091I9B8 A0A091U0A3 A0A087XVL0 G3TXX8 M3ZZ73 M3Z0Q0 U6D258 K9J019 A0A250XYX9 A0A2I2V251 A0A091L3V4 A0A3P9Q8Z3 A0A093Q836 A0A091RRN5 G1LSL4 A0A452E3U5 A0A091KS60 D1ZZC7 A0A0A0AT93 G1PWI9 L5K8L5 A0A3Q2HSD8 A0A091NRN2 W5N9J1 A0A2K6DFS0 A0A2I3FW14 A0A3P4RUE5 A0A091UMD1 A0A452E3S8 A0A093H5Q6 A0A2K5L7T3 F7D6G8 Q9GKU5 A0A452E3T6 A0A093R3X4 A0A091RKV4 G1QY87 A0A087VE56 G7NYG2 G7MKN7 A0A091MIM1 A0A091J365 A0A1S3AA44 A0A091WJ64
A0A067R947 A0A2J7PGD8 A0A2P8XLX9 I3LTU6 A0A1B6MAN5 A0A2Y9ENA6 A0A1Y1LWP4 A0A1B6DMB6 A0A384A6S9 A0A341CY01 G3SLU4 A0A2Y9MM77 A0A099ZPM0 A0A093I875 A0A2K6U715 A0A091EX69 A0A3Q7Q521 A0A2Y9HGJ3 A0A2K6U773 A0A2Y9D5Z2 K7DAK7 A0A024R2I1 Q96DB2 A0A2K5SAC0 G3QXL9 A0A3Q2KRU9 F6WJS2 A0A2J8NNX6 A0A2J8TVT1 A0A2K5JRW1 A0A2K6QLH9 A0A2K6KMM0 F7HYS2 A0A3Q2I1P4 A0A2K5ETR4 A0A452QXR8 A0A2K5JRT8 A0A3B3WIE7 A0A091TKR1 A0A093CY31 A0A093BLT0 A0A2J8TVT9 A0A3Q7V7T1 A0A2U3VLA8 H2P9B0 A0A3Q7RNE1 A0A091Q2D1 A0A402EFM4 M3W7T9 A0A2I2UGE1 A0A091I9B8 A0A091U0A3 A0A087XVL0 G3TXX8 M3ZZ73 M3Z0Q0 U6D258 K9J019 A0A250XYX9 A0A2I2V251 A0A091L3V4 A0A3P9Q8Z3 A0A093Q836 A0A091RRN5 G1LSL4 A0A452E3U5 A0A091KS60 D1ZZC7 A0A0A0AT93 G1PWI9 L5K8L5 A0A3Q2HSD8 A0A091NRN2 W5N9J1 A0A2K6DFS0 A0A2I3FW14 A0A3P4RUE5 A0A091UMD1 A0A452E3S8 A0A093H5Q6 A0A2K5L7T3 F7D6G8 Q9GKU5 A0A452E3T6 A0A093R3X4 A0A091RKV4 G1QY87 A0A087VE56 G7NYG2 G7MKN7 A0A091MIM1 A0A091J365 A0A1S3AA44 A0A091WJ64
EC Number
3.5.1.98
Pubmed
EMBL
BABH01032915
BABH01032916
BABH01032917
RSAL01000059
RVE49681.1
ODYU01003513
+ More
SOQ42398.1 KQ459460 KPJ00680.1 KQ459986 KPJ18824.1 GECZ01024746 JAS45023.1 KK852614 KDR20203.1 NEVH01025635 PNF15398.1 PYGN01001751 PSN32998.1 AEMK02000087 DQIR01212448 HDB67925.1 GEBQ01019799 GEBQ01006988 JAT20178.1 JAT32989.1 GEZM01050646 GEZM01050644 GEZM01050643 GEZM01050639 JAV75447.1 GEDC01025617 GEDC01010538 JAS11681.1 JAS26760.1 KL897327 KGL84344.1 KL206989 KFV88095.1 KK719041 KFO60924.1 AACZ04000097 AACZ04000098 AACZ04000099 GABC01003692 GABF01002886 GABD01010863 GABE01007395 NBAG03000225 JAA07646.1 JAA19259.1 JAA22237.1 JAA37344.1 PNI73437.1 CH471055 EAW64168.1 AK025426 AK025890 AK293223 AC027124 BC009676 AL137362 CABD030019961 PNI73447.1 NDHI03003481 PNJ37136.1 GAMT01004432 GAMS01009080 GAMR01009255 GAMQ01006595 GAMP01001054 JAB07429.1 JAB14056.1 JAB24677.1 JAB35256.1 JAB51701.1 KK455221 KFQ75900.1 KL472461 KFV19388.1 KN125955 KFU85335.1 PNJ37144.1 ABGA01348650 ABGA01348651 ABGA01348652 KK683734 KFQ13706.1 BDOT01000009 GCF42970.1 AANG04003845 KL218426 KFP04822.1 KK408647 KFQ83482.1 AYCK01015304 AEYP01003192 HAAF01004824 CCP76649.1 GABZ01006831 JAA46694.1 GFFV01003348 JAV36597.1 KL300137 KFP51139.1 KL672103 KFW84731.1 KK929451 KFQ44466.1 ACTA01016553 LWLT01000024 KK756336 KFP43459.1 KQ971338 EFA01868.1 KL872890 KGL97181.1 AAPE02010885 KB030947 ELK07712.1 KL391482 KFP91747.1 AHAT01015107 AHAT01015108 ADFV01026286 ADFV01026287 ADFV01026288 ADFV01026289 ADFV01026290 ADFV01026291 CYRY02043874 VCX38544.1 KL409829 KFQ92099.1 KK618797 KFV50053.1 JSUE03023159 JV636381 AFJ71721.1 AB052134 KL224925 KFW65714.1 KK819795 KFQ39614.1 KL489887 KFO10898.1 CM001277 EHH51121.1 CM001254 EHH16157.1 KK828903 KFP74908.1 KK501311 KFP14150.1 KK735759 KFR15727.1
SOQ42398.1 KQ459460 KPJ00680.1 KQ459986 KPJ18824.1 GECZ01024746 JAS45023.1 KK852614 KDR20203.1 NEVH01025635 PNF15398.1 PYGN01001751 PSN32998.1 AEMK02000087 DQIR01212448 HDB67925.1 GEBQ01019799 GEBQ01006988 JAT20178.1 JAT32989.1 GEZM01050646 GEZM01050644 GEZM01050643 GEZM01050639 JAV75447.1 GEDC01025617 GEDC01010538 JAS11681.1 JAS26760.1 KL897327 KGL84344.1 KL206989 KFV88095.1 KK719041 KFO60924.1 AACZ04000097 AACZ04000098 AACZ04000099 GABC01003692 GABF01002886 GABD01010863 GABE01007395 NBAG03000225 JAA07646.1 JAA19259.1 JAA22237.1 JAA37344.1 PNI73437.1 CH471055 EAW64168.1 AK025426 AK025890 AK293223 AC027124 BC009676 AL137362 CABD030019961 PNI73447.1 NDHI03003481 PNJ37136.1 GAMT01004432 GAMS01009080 GAMR01009255 GAMQ01006595 GAMP01001054 JAB07429.1 JAB14056.1 JAB24677.1 JAB35256.1 JAB51701.1 KK455221 KFQ75900.1 KL472461 KFV19388.1 KN125955 KFU85335.1 PNJ37144.1 ABGA01348650 ABGA01348651 ABGA01348652 KK683734 KFQ13706.1 BDOT01000009 GCF42970.1 AANG04003845 KL218426 KFP04822.1 KK408647 KFQ83482.1 AYCK01015304 AEYP01003192 HAAF01004824 CCP76649.1 GABZ01006831 JAA46694.1 GFFV01003348 JAV36597.1 KL300137 KFP51139.1 KL672103 KFW84731.1 KK929451 KFQ44466.1 ACTA01016553 LWLT01000024 KK756336 KFP43459.1 KQ971338 EFA01868.1 KL872890 KGL97181.1 AAPE02010885 KB030947 ELK07712.1 KL391482 KFP91747.1 AHAT01015107 AHAT01015108 ADFV01026286 ADFV01026287 ADFV01026288 ADFV01026289 ADFV01026290 ADFV01026291 CYRY02043874 VCX38544.1 KL409829 KFQ92099.1 KK618797 KFV50053.1 JSUE03023159 JV636381 AFJ71721.1 AB052134 KL224925 KFW65714.1 KK819795 KFQ39614.1 KL489887 KFO10898.1 CM001277 EHH51121.1 CM001254 EHH16157.1 KK828903 KFP74908.1 KK501311 KFP14150.1 KK735759 KFR15727.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000053240
UP000027135
UP000235965
+ More
UP000245037 UP000008227 UP000248484 UP000261681 UP000252040 UP000007646 UP000248483 UP000053641 UP000053584 UP000233220 UP000052976 UP000286641 UP000248481 UP000248480 UP000002277 UP000005640 UP000233040 UP000001519 UP000002281 UP000233080 UP000233200 UP000233180 UP000008225 UP000233020 UP000291022 UP000261480 UP000286642 UP000245340 UP000001595 UP000286640 UP000288954 UP000011712 UP000054308 UP000028760 UP000002852 UP000000715 UP000242638 UP000053258 UP000008912 UP000291000 UP000007266 UP000053858 UP000001074 UP000010552 UP000018468 UP000233120 UP000001073 UP000053283 UP000233060 UP000006718 UP000233100 UP000054081 UP000009130 UP000053119 UP000079721 UP000053605
UP000245037 UP000008227 UP000248484 UP000261681 UP000252040 UP000007646 UP000248483 UP000053641 UP000053584 UP000233220 UP000052976 UP000286641 UP000248481 UP000248480 UP000002277 UP000005640 UP000233040 UP000001519 UP000002281 UP000233080 UP000233200 UP000233180 UP000008225 UP000233020 UP000291022 UP000261480 UP000286642 UP000245340 UP000001595 UP000286640 UP000288954 UP000011712 UP000054308 UP000028760 UP000002852 UP000000715 UP000242638 UP000053258 UP000008912 UP000291000 UP000007266 UP000053858 UP000001074 UP000010552 UP000018468 UP000233120 UP000001073 UP000053283 UP000233060 UP000006718 UP000233100 UP000054081 UP000009130 UP000053119 UP000079721 UP000053605
Pfam
PF00850 Hist_deacetyl
Interpro
SUPFAM
SSF52768
SSF52768
Gene 3D
ProteinModelPortal
H9JIP7
A0A3S2TLX4
A0A2H1VNL7
A0A194Q6P7
A0A0N1PHA1
A0A1B6F569
+ More
A0A067R947 A0A2J7PGD8 A0A2P8XLX9 I3LTU6 A0A1B6MAN5 A0A2Y9ENA6 A0A1Y1LWP4 A0A1B6DMB6 A0A384A6S9 A0A341CY01 G3SLU4 A0A2Y9MM77 A0A099ZPM0 A0A093I875 A0A2K6U715 A0A091EX69 A0A3Q7Q521 A0A2Y9HGJ3 A0A2K6U773 A0A2Y9D5Z2 K7DAK7 A0A024R2I1 Q96DB2 A0A2K5SAC0 G3QXL9 A0A3Q2KRU9 F6WJS2 A0A2J8NNX6 A0A2J8TVT1 A0A2K5JRW1 A0A2K6QLH9 A0A2K6KMM0 F7HYS2 A0A3Q2I1P4 A0A2K5ETR4 A0A452QXR8 A0A2K5JRT8 A0A3B3WIE7 A0A091TKR1 A0A093CY31 A0A093BLT0 A0A2J8TVT9 A0A3Q7V7T1 A0A2U3VLA8 H2P9B0 A0A3Q7RNE1 A0A091Q2D1 A0A402EFM4 M3W7T9 A0A2I2UGE1 A0A091I9B8 A0A091U0A3 A0A087XVL0 G3TXX8 M3ZZ73 M3Z0Q0 U6D258 K9J019 A0A250XYX9 A0A2I2V251 A0A091L3V4 A0A3P9Q8Z3 A0A093Q836 A0A091RRN5 G1LSL4 A0A452E3U5 A0A091KS60 D1ZZC7 A0A0A0AT93 G1PWI9 L5K8L5 A0A3Q2HSD8 A0A091NRN2 W5N9J1 A0A2K6DFS0 A0A2I3FW14 A0A3P4RUE5 A0A091UMD1 A0A452E3S8 A0A093H5Q6 A0A2K5L7T3 F7D6G8 Q9GKU5 A0A452E3T6 A0A093R3X4 A0A091RKV4 G1QY87 A0A087VE56 G7NYG2 G7MKN7 A0A091MIM1 A0A091J365 A0A1S3AA44 A0A091WJ64
A0A067R947 A0A2J7PGD8 A0A2P8XLX9 I3LTU6 A0A1B6MAN5 A0A2Y9ENA6 A0A1Y1LWP4 A0A1B6DMB6 A0A384A6S9 A0A341CY01 G3SLU4 A0A2Y9MM77 A0A099ZPM0 A0A093I875 A0A2K6U715 A0A091EX69 A0A3Q7Q521 A0A2Y9HGJ3 A0A2K6U773 A0A2Y9D5Z2 K7DAK7 A0A024R2I1 Q96DB2 A0A2K5SAC0 G3QXL9 A0A3Q2KRU9 F6WJS2 A0A2J8NNX6 A0A2J8TVT1 A0A2K5JRW1 A0A2K6QLH9 A0A2K6KMM0 F7HYS2 A0A3Q2I1P4 A0A2K5ETR4 A0A452QXR8 A0A2K5JRT8 A0A3B3WIE7 A0A091TKR1 A0A093CY31 A0A093BLT0 A0A2J8TVT9 A0A3Q7V7T1 A0A2U3VLA8 H2P9B0 A0A3Q7RNE1 A0A091Q2D1 A0A402EFM4 M3W7T9 A0A2I2UGE1 A0A091I9B8 A0A091U0A3 A0A087XVL0 G3TXX8 M3ZZ73 M3Z0Q0 U6D258 K9J019 A0A250XYX9 A0A2I2V251 A0A091L3V4 A0A3P9Q8Z3 A0A093Q836 A0A091RRN5 G1LSL4 A0A452E3U5 A0A091KS60 D1ZZC7 A0A0A0AT93 G1PWI9 L5K8L5 A0A3Q2HSD8 A0A091NRN2 W5N9J1 A0A2K6DFS0 A0A2I3FW14 A0A3P4RUE5 A0A091UMD1 A0A452E3S8 A0A093H5Q6 A0A2K5L7T3 F7D6G8 Q9GKU5 A0A452E3T6 A0A093R3X4 A0A091RKV4 G1QY87 A0A087VE56 G7NYG2 G7MKN7 A0A091MIM1 A0A091J365 A0A1S3AA44 A0A091WJ64
PDB
1C3P
E-value=4.34604e-14,
Score=188
Ontologies
GO
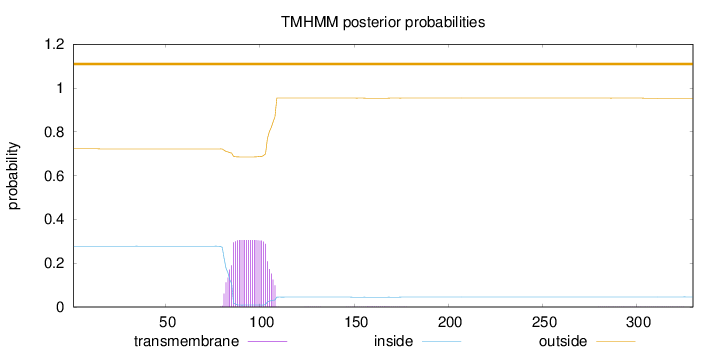
Topology
Subcellular location
Nucleus
Length:
330
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
6.95041
Exp number, first 60 AAs:
0.00189
Total prob of N-in:
0.27775
outside
1 - 330
Population Genetic Test Statistics
Pi
223.820715
Theta
187.938993
Tajima's D
0.779426
CLR
0.004584
CSRT
0.588820558972051
Interpretation
Uncertain
