Gene
KWMTBOMO08445 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009402
Annotation
MESH_BOMMO_RecName:_Full%3DProtein_mesh
Full name
Protein mesh
Location in the cell
PlasmaMembrane Reliability : 1.922
Sequence
CDS
ATGGGCTCGACGACCTACCAGAACAATGGACAGCCATACGTCATCACCACCCAGCGCCTGCAGCAGATCCGCTCCAATTTTATGTACTGGTTCTACGATCAAGGAGGCAGCGATAACATCGGTGACTACCAGAGAGACATACACACTTCCACGCCGCAGATTCACAAGAATTTCAACTTTCAACTGCCATTCTTTGGGTTCAGATTCAATTACACAAGGATCTCGATGAATGGGTACATTTACTTCAGTGACCCCCCAGACCACTACACCTACCCATTGTCGTTTCCCGTGCGAGACTGGCCGAATATCAACGATCCTTCTTTCATAGGGATATTCTTCAGCAAGTGTCGTATTGGTAACATGAGACCCGAAGAGCCTGATCCGAGAAGACCTGGAATATACTTCAGATTGGACAGAGACTTACAGACACGTACGGACCAACTGGGAGTCGAGATGCGAGAACGCGTGACGTGGGACATCCGTGAGGGCGTCATCGGCTCTGAGACCTTCTTCCCGAAACATACAATAACGATCACGTGGAAGAACATGTCGTTTGCCGGCGGTATTGACAACTCGCTGTTTATGACAAACACATTCCAAATGGTTCTAGCAACGGATGAAGTCTTCACTTATGCGATTTTCAATTATCTCGAAATAAATTGGAGTTCTCACACTGAAGCTGGCGGAGACACCACTACCGGTGAAGGAGGCATACCGGCCTATATTGGCTTCAACGCTGGTAACGGGACACGAAGTTACGAATACAAACCGTATTCTCAAGCTTCTGTGCTTCGAGATTTGACCGGCAGAGGCTGGGCCAATGGCTTTCCAGGGCGACACATATTCAGAATTGACGAGAATATACTAATGGGCACTTGTAATAAGGATATCGACGGCGCCAACCTGCCCCTGATGTTTGCGCCAGAGAGTGGTAACATGCTCGGCGGCACAATCGTCAATATCACGGGTCCATGCTTCAATCCCAACGATAGAATAACGTGCAGATTCGATACAGAATCCGTTCTCGGAGCTGTTGTGGACGTCAACAGGGCTATCTGTGTGCAACCAAGATTCTGGCACAATGGTTACGCAAGATTTGAAGTTGCCATTAACAATGAACCTTATAAATGGAAAGGAAGATATTTTGTTGAAACCCCTGCGACCGCAACCGAAAAGATATTTTTCCCAGATAATTCTGTACACGAAAGATATCCCCCAGAAGTCCGAATAACTTGGGACCGATTTAATCTGACAACAAACCTAAACGTTCAGCTCCAAATCAGTTTGTGGGGCTACAAAGAAGTCACGATCCGACCTCAATTAGAATACATCGATATGATAGAAGTCGGAGTTGCGAACACCGGTGAATACGTGATAAATCCACAAAATTTCCGGAACAGGGAAAACATCATGCACAACGACATGCAATTCGGTTTTCTGCAAATCAATTTGACCACTCCCGAAGTATTCAAAGGGGTTCCTATATCTCCGATTCTATGGAGTCGTCCGATCCCACTAGGTTGGTATTTCGCCCCACAGTGGGAAAGGCTTCACGGACAGCGCTGGTCGAACTCCATGTGTAACAACTGGCTTCGAACCGATCGTTTCCTTAAGAACTTTGCAGCTCAAGTTTGGGTCTGCCCCTGTACTTTGGAACACGCGCTTCTTGATAAAGGTCGCTTCATGCCGGACTTGGACTGCGACAGAGACACGAACCCTACTTGTAGATATCATTGGGGTGGTATTCATTGCGTCAGGAGTGGTGCTCCAAGTTCGGAAGGTTCTGGCCAGCAATGCTGCTACGACAAAAACGGCTTCCTCATGTTATCCTACGACCAGATGTGGGGCTCAAAGCCGTCAAGATCTCACGATTTCGGATTTACACCTTACAACGAAGCCAATAAGGTTCCGTCGTTATCTCGCTGGTTCCACGACATGATTCCGTTCTATCAATGTTGTTTGTGGCAAGAAGAACAAGCTGTCGGTTGCGAGACTTTCAGGTTTGAACGTCGACCGTCTCAAGACTGCGTGGCCTATCAATCTCCAGGAGTCGCGGGTATCTTCGGTGATCCACATATAGTCACCTTTGACGACCTGCAGTACACTTTCAATGGCAAAGGTGAATACGTCCTAGTGAGGGTGGATCATTCACAACTCAAGCTCGACGTTCAAGGTAGATTTGAACAGGTACCAAGAAATATTCACGGTGCGGTCAACGCGACACACTTAACGTCAGTCGTAGCAGCGTCCAACAACTCTCAAACCATTGAGGTACGCCTTCGACCACAACACGCTCAGTGGAGGTACAGGCTCGATGTTTTCGCCAATGGCAAGAGAGTCTACTTCGACAGGACTGCACTAAGAGTACAATATTTCCCAGGTGTCACCGTGTACCAGCCTATGTACGTCCTAAACCAGTCGGAAATCGTCGTTATGTTCTCGTCTGGGGCCGGATTGGAAGTAGTCGAGAATCGCGGCTTCATGACTGCGAGAGTTTATTTACCATGGACGTTTATGAATCAAACTCGAGGATTATTCGGCAACTGGTCTCTGGATGTGAACGATGACTTCACAAGACCCGATGGCACGCTGGCATCGGTCGATCTGAACAATTTCCAATCCGCCCACAGAGATTTTGCACAGCACTGGCAACTCACAGATCGTGAGCAACGGGACATCGGCGTGGCGATGTTCGTTAGAGAATATGGTCGGACGGCGGCTTATTACAACGACAATGAATTTATACCGAACTTCATACGAGAACCGGCGAACTTCTTGCCGGTGAACCGTTCTCACGACGTGACGAGAGCTATCGAGATATGCCAAGATTCTTATCAGTGTCGATACGATTACGGCATGACCCTCAATCGGGATATGGCTGAATTTACGAAGAATTATCTCTCATCGATTACTAATATAAAAGAGCAGAACGCAAGGCGGGTCATCAGTTGCGGTATTCTGGAGACGCCACGTTTTGGAAGAAAGAGTAACTTCTTCTTCACGCCTGGAACCAGGGTAAACTTCGAATGTAATCAAGACTTCATTCTGACCGGGGACAAAAGACGAGTGTGTGAAGACAACGGCAGATGGAATCTCCCTGACTATGGATACACAGAGTGTCTACGTCAACAGGAATTCTCCCAACGTGCCCTATTCCTAACTTGGGGCGTCATAGTGGCAGTTATTCTACCATTAGGCCTCTTGATTTGCCTCCTCTGGTTCTGGTGCTGGCACAAACCAAGGTCCGAGGGCAAGGAGGGCTTCCGTTTCGAAGACTTACCGCGTTCAAAATCCGCATCTAGATTAAATCTTAGGTCTTCGTCAATGGGAAATATCACAGACACTATGAAATCTTCGACGATACCCGGTTCTGAGAAAAAATCCCCCGAAACCCCTACTGAGGAAACCCCGGCCAGGATAGTAGGCAGGTCAGTCCTCGCTCCTCCCGCGGACGGAGACAGCTCAGGAATAGGCTACCCAGACTCTGGCAAAAGTGATTCCGGTAAATCTGACAAGTCCTCCGGTCTGCCTAAAAAGCGTAGGGCTTACGATAAAACATATCGTACCAATGAGCCATTACCGAATGCACCCGATGTAGAATTCCCAGAAAAACTATGGGATCTGTCCGAGGAAGACTTGTTGTCACTGACTTCACCATCAGATTCTGAATCTAATAGAGATTCCACTCTAACTCGTCCGGCTAAAGACATACAGTATCTAAACAAACCGCGACAAACCGGTCGTCAGGCCATACCGTCCGACTCAGGCTATTCCACGAAGGAGGGATCTGAGGATCCTTACGCGCCGAAATTCGACGACCAATACAGTCCAATACCTTCTCAGTATTCTCCCACGTACTCTGAAATTTATTCGCCTCCAATCAGCCCGGCCTCCGATTCGAGTCCCCGGAATACATATAATAATCCCGGCATTCCCGAAGCACCTAAAAGCGCCCCTGTTGATGGCATTAAAACTTTTACCATGCCAACGAACAAGGGTAAACAGGAGTACTCGTCTCGGACTCTTGGTGCGACATGGGGCATCATTAGTGCTGTGATGCTGCCAATTATAATAATCTTAATCTGCGTCGCTTGGAGGATCTTGCAGAGGCGGAAAGCTGAAGAAAGAGAAGAAAACGAATTCTTGGATGTGAAGACCCGTGCAATCGATCCTGACGATTCAGTTAAAGTGACTTCAGACGATGAAAGCATTCCTTACAAGAAAGACGTAACAGAAGAGACGCCTGAGCCCACAGAAGGCGTCCAGGCGGTGGAACCCTCCAATCCGAACTACAATTACGGTCGCCCCTACGTTGACCTCCAACCGGGACAACCGAGGCAATGGGGTGGAGAAACTGAAATTAATTAA
Protein
MGSTTYQNNGQPYVITTQRLQQIRSNFMYWFYDQGGSDNIGDYQRDIHTSTPQIHKNFNFQLPFFGFRFNYTRISMNGYIYFSDPPDHYTYPLSFPVRDWPNINDPSFIGIFFSKCRIGNMRPEEPDPRRPGIYFRLDRDLQTRTDQLGVEMRERVTWDIREGVIGSETFFPKHTITITWKNMSFAGGIDNSLFMTNTFQMVLATDEVFTYAIFNYLEINWSSHTEAGGDTTTGEGGIPAYIGFNAGNGTRSYEYKPYSQASVLRDLTGRGWANGFPGRHIFRIDENILMGTCNKDIDGANLPLMFAPESGNMLGGTIVNITGPCFNPNDRITCRFDTESVLGAVVDVNRAICVQPRFWHNGYARFEVAINNEPYKWKGRYFVETPATATEKIFFPDNSVHERYPPEVRITWDRFNLTTNLNVQLQISLWGYKEVTIRPQLEYIDMIEVGVANTGEYVINPQNFRNRENIMHNDMQFGFLQINLTTPEVFKGVPISPILWSRPIPLGWYFAPQWERLHGQRWSNSMCNNWLRTDRFLKNFAAQVWVCPCTLEHALLDKGRFMPDLDCDRDTNPTCRYHWGGIHCVRSGAPSSEGSGQQCCYDKNGFLMLSYDQMWGSKPSRSHDFGFTPYNEANKVPSLSRWFHDMIPFYQCCLWQEEQAVGCETFRFERRPSQDCVAYQSPGVAGIFGDPHIVTFDDLQYTFNGKGEYVLVRVDHSQLKLDVQGRFEQVPRNIHGAVNATHLTSVVAASNNSQTIEVRLRPQHAQWRYRLDVFANGKRVYFDRTALRVQYFPGVTVYQPMYVLNQSEIVVMFSSGAGLEVVENRGFMTARVYLPWTFMNQTRGLFGNWSLDVNDDFTRPDGTLASVDLNNFQSAHRDFAQHWQLTDREQRDIGVAMFVREYGRTAAYYNDNEFIPNFIREPANFLPVNRSHDVTRAIEICQDSYQCRYDYGMTLNRDMAEFTKNYLSSITNIKEQNARRVISCGILETPRFGRKSNFFFTPGTRVNFECNQDFILTGDKRRVCEDNGRWNLPDYGYTECLRQQEFSQRALFLTWGVIVAVILPLGLLICLLWFWCWHKPRSEGKEGFRFEDLPRSKSASRLNLRSSSMGNITDTMKSSTIPGSEKKSPETPTEETPARIVGRSVLAPPADGDSSGIGYPDSGKSDSGKSDKSSGLPKKRRAYDKTYRTNEPLPNAPDVEFPEKLWDLSEEDLLSLTSPSDSESNRDSTLTRPAKDIQYLNKPRQTGRQAIPSDSGYSTKEGSEDPYAPKFDDQYSPIPSQYSPTYSEIYSPPISPASDSSPRNTYNNPGIPEAPKSAPVDGIKTFTMPTNKGKQEYSSRTLGATWGIISAVMLPIIIILICVAWRILQRRKAEEREENEFLDVKTRAIDPDDSVKVTSDDESIPYKKDVTEETPEPTEGVQAVEPSNPNYNYGRPYVDLQPGQPRQWGGETEIN
Summary
Description
May be required for the proper organization of smooth septate junctions and for the barrier function of the midgut epithelium.
Required, together with Ssk and Tsp2A, for the proper organization of smooth septate junctions (sSJs), probably by mediating cell adhesion via its homophilic interaction (PubMed:22854041, PubMed:26848177). Also required for the correct subcellular localization of several sSJ components, such as Ssk, cora and l(2)gl, and for the barrier function of the midgut epithelium (PubMed:22854041). Required for maintaining the three-layered structure of the proventriculus (PubMed:22854041).
Required, together with Ssk and Tsp2A, for the proper organization of smooth septate junctions (sSJs), probably by mediating cell adhesion via its homophilic interaction (PubMed:22854041, PubMed:26848177). Also required for the correct subcellular localization of several sSJ components, such as Ssk, cora and l(2)gl, and for the barrier function of the midgut epithelium (PubMed:22854041). Required for maintaining the three-layered structure of the proventriculus (PubMed:22854041).
Subunit
Forms a complex with Ssk and Tsp2A (PubMed:26848177). Interacts with Ssk; the interaction may be necessary for the localization of both proteins to the cell apicolateral region (PubMed:22854041).
Keywords
Cell junction
Cell membrane
Complete proteome
Disulfide bond
Membrane
Phosphoprotein
Reference proteome
Signal
Sushi
Transmembrane
Transmembrane helix
Alternative splicing
Cell adhesion
Developmental protein
Glycoprotein
Feature
chain Protein mesh
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9JIQ1
A0A0N1IBZ5
A0A0N0PAJ1
A0A2A4JZQ1
A0A212FC21
A0A1E1WKK5
+ More
A0A1E1WUZ1 A0A2A4K0U0 A0A1S4FAM4 A0A1S4FAU2 A0A1L8DPS8 A0A1L8DPH4 Q17A21 Q17A20 A0A182G1X8 A0A0L7KX63 K7J8G5 A0A1Q3FIW5 A0A1Q3FJ17 A0A1Q3FJ00 A0A1Q3FJ26 A0A084VQK1 A0A336M1P2 A0A182P7W7 B0XCS0 A0A182IA85 A0A182JSU7 A0A182UY80 Q7QFL4 A0A182FIZ9 A0A182QHW0 A0A1S4G989 A0A182W4P0 A0A182JG23 A0A1J1HSW6 A0A182MJE9 A0A182RBH7 W5JLJ6 A0A1J1HSV2 A0A336M555 A0A0M9A432 A0A1W4X666 A0A1W4XFP3 A0A1W4XGL2 A0A2H8TDI6 J9JV68 A0A2S2QM76 A0A182Y7J4 A0A0C9QJR1 A0A151I9C3 A0A182N115 A0A1B6DF11 A0A1B6ECQ7 A0A2A3ETZ4 A0A0L0BY61 E2AP22 A0A088AMW9 F4X0V1 A0A151HZ41 A0A158N9E9 E2BB29 A0A1Y1KJZ8 A0A026WGR5 A0A067RID9 A0A1A9WZ27 T1PGP1 A0A195E220 A0A151JV66 A0A1L8EAD8 E0VDP6 A0A1I8P1Y8 A0A224XA01 A0A1I8P260 A0A1L8EHL7 A0A1B0FP21 T1PEZ0 A0A151X2L5 A0A1Y1KK87 T1PEU8 T1PHE4 A0A1L8EAN0 A0A034UZJ8 A0A1L8EA36 B4K6G9 A0A1W4U686 A0A1I8M7B4 A0A1W4UHW4 A0A1W4UHZ5 A0A3B0JKH5 B4HZZ6 A0A3B0JEL6 A0A0K8WM51 B3P7S3 B4M5V3 A0A3B0JJ83 Q0KHY3 Q0KHY3-1 I5AMT6
A0A1E1WUZ1 A0A2A4K0U0 A0A1S4FAM4 A0A1S4FAU2 A0A1L8DPS8 A0A1L8DPH4 Q17A21 Q17A20 A0A182G1X8 A0A0L7KX63 K7J8G5 A0A1Q3FIW5 A0A1Q3FJ17 A0A1Q3FJ00 A0A1Q3FJ26 A0A084VQK1 A0A336M1P2 A0A182P7W7 B0XCS0 A0A182IA85 A0A182JSU7 A0A182UY80 Q7QFL4 A0A182FIZ9 A0A182QHW0 A0A1S4G989 A0A182W4P0 A0A182JG23 A0A1J1HSW6 A0A182MJE9 A0A182RBH7 W5JLJ6 A0A1J1HSV2 A0A336M555 A0A0M9A432 A0A1W4X666 A0A1W4XFP3 A0A1W4XGL2 A0A2H8TDI6 J9JV68 A0A2S2QM76 A0A182Y7J4 A0A0C9QJR1 A0A151I9C3 A0A182N115 A0A1B6DF11 A0A1B6ECQ7 A0A2A3ETZ4 A0A0L0BY61 E2AP22 A0A088AMW9 F4X0V1 A0A151HZ41 A0A158N9E9 E2BB29 A0A1Y1KJZ8 A0A026WGR5 A0A067RID9 A0A1A9WZ27 T1PGP1 A0A195E220 A0A151JV66 A0A1L8EAD8 E0VDP6 A0A1I8P1Y8 A0A224XA01 A0A1I8P260 A0A1L8EHL7 A0A1B0FP21 T1PEZ0 A0A151X2L5 A0A1Y1KK87 T1PEU8 T1PHE4 A0A1L8EAN0 A0A034UZJ8 A0A1L8EA36 B4K6G9 A0A1W4U686 A0A1I8M7B4 A0A1W4UHW4 A0A1W4UHZ5 A0A3B0JKH5 B4HZZ6 A0A3B0JEL6 A0A0K8WM51 B3P7S3 B4M5V3 A0A3B0JJ83 Q0KHY3 Q0KHY3-1 I5AMT6
Pubmed
EMBL
KQ459986
KPJ18828.1
KQ458751
KPJ05298.1
NWSH01000337
PCG77289.1
+ More
AGBW02009237 OWR51289.1 GDQN01003663 JAT87391.1 GDQN01000270 JAT90784.1 PCG77290.1 GFDF01005729 JAV08355.1 GFDF01005728 JAV08356.1 CH477341 EAT43085.1 EAT43086.1 JXUM01039553 KQ561209 KXJ79338.1 JTDY01004823 KOB67705.1 GFDL01007495 JAV27550.1 GFDL01007527 JAV27518.1 GFDL01007478 JAV27567.1 GFDL01007517 JAV27528.1 ATLV01015219 KE525003 KFB40245.1 UFQT01000418 SSX24154.1 DS232716 EDS45087.1 APCN01001132 AAAB01008846 EAA06289.5 AXCN02001386 CVRI01000020 CRK91151.1 AXCM01001653 ADMH02001211 ETN63649.1 CRK91152.1 UFQT01000413 SSX24083.1 KQ435740 KOX76807.1 GFXV01000325 MBW12130.1 ABLF02029367 ABLF02029368 GGMS01009437 MBY78640.1 GBYB01015053 JAG84820.1 KQ978285 KYM95672.1 GEDC01013076 JAS24222.1 GEDC01013156 GEDC01001644 JAS24142.1 JAS35654.1 KZ288190 PBC34521.1 JRES01001160 KNC24960.1 GL441439 EFN64854.1 GL888503 EGI59893.1 KQ976712 KYM77000.1 ADTU01009566 GL446901 EFN87120.1 GEZM01081254 JAV61733.1 KK107214 QOIP01000005 EZA55257.1 RLU22395.1 KK852614 KDR20197.1 KA647310 AFP61939.1 KQ979814 KYN18949.1 KQ981740 KYN36512.1 GFDG01003175 JAV15624.1 DS235083 EEB11502.1 GFTR01008692 JAW07734.1 GFDG01000592 JAV18207.1 CCAG010007645 CCAG010007646 KA647311 AFP61940.1 KQ982572 KYQ54687.1 GEZM01081253 JAV61734.1 KA647312 AFP61941.1 KA647313 AFP61942.1 GFDG01003176 JAV15623.1 GAKP01022511 JAC36441.1 GFDG01003202 JAV15597.1 CH933806 EDW16269.2 OUUW01000005 SPP80822.1 CH480819 EDW53603.1 SPP80824.1 GDHF01000101 JAI52213.1 CH954182 EDV52981.1 CH940652 EDW59029.2 SPP80823.1 AE014297 BT032891 BT133164 BT011360 AY122109 CM000070 EIM52271.1
AGBW02009237 OWR51289.1 GDQN01003663 JAT87391.1 GDQN01000270 JAT90784.1 PCG77290.1 GFDF01005729 JAV08355.1 GFDF01005728 JAV08356.1 CH477341 EAT43085.1 EAT43086.1 JXUM01039553 KQ561209 KXJ79338.1 JTDY01004823 KOB67705.1 GFDL01007495 JAV27550.1 GFDL01007527 JAV27518.1 GFDL01007478 JAV27567.1 GFDL01007517 JAV27528.1 ATLV01015219 KE525003 KFB40245.1 UFQT01000418 SSX24154.1 DS232716 EDS45087.1 APCN01001132 AAAB01008846 EAA06289.5 AXCN02001386 CVRI01000020 CRK91151.1 AXCM01001653 ADMH02001211 ETN63649.1 CRK91152.1 UFQT01000413 SSX24083.1 KQ435740 KOX76807.1 GFXV01000325 MBW12130.1 ABLF02029367 ABLF02029368 GGMS01009437 MBY78640.1 GBYB01015053 JAG84820.1 KQ978285 KYM95672.1 GEDC01013076 JAS24222.1 GEDC01013156 GEDC01001644 JAS24142.1 JAS35654.1 KZ288190 PBC34521.1 JRES01001160 KNC24960.1 GL441439 EFN64854.1 GL888503 EGI59893.1 KQ976712 KYM77000.1 ADTU01009566 GL446901 EFN87120.1 GEZM01081254 JAV61733.1 KK107214 QOIP01000005 EZA55257.1 RLU22395.1 KK852614 KDR20197.1 KA647310 AFP61939.1 KQ979814 KYN18949.1 KQ981740 KYN36512.1 GFDG01003175 JAV15624.1 DS235083 EEB11502.1 GFTR01008692 JAW07734.1 GFDG01000592 JAV18207.1 CCAG010007645 CCAG010007646 KA647311 AFP61940.1 KQ982572 KYQ54687.1 GEZM01081253 JAV61734.1 KA647312 AFP61941.1 KA647313 AFP61942.1 GFDG01003176 JAV15623.1 GAKP01022511 JAC36441.1 GFDG01003202 JAV15597.1 CH933806 EDW16269.2 OUUW01000005 SPP80822.1 CH480819 EDW53603.1 SPP80824.1 GDHF01000101 JAI52213.1 CH954182 EDV52981.1 CH940652 EDW59029.2 SPP80823.1 AE014297 BT032891 BT133164 BT011360 AY122109 CM000070 EIM52271.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000007151
UP000008820
+ More
UP000069940 UP000249989 UP000037510 UP000002358 UP000030765 UP000075885 UP000002320 UP000075840 UP000075881 UP000075903 UP000007062 UP000069272 UP000075886 UP000075920 UP000075880 UP000183832 UP000075883 UP000075900 UP000000673 UP000053105 UP000192223 UP000007819 UP000076408 UP000078542 UP000075884 UP000242457 UP000037069 UP000000311 UP000005203 UP000007755 UP000078540 UP000005205 UP000008237 UP000053097 UP000279307 UP000027135 UP000091820 UP000078492 UP000078541 UP000009046 UP000095300 UP000092444 UP000075809 UP000009192 UP000192221 UP000095301 UP000268350 UP000001292 UP000008711 UP000008792 UP000000803 UP000001819
UP000069940 UP000249989 UP000037510 UP000002358 UP000030765 UP000075885 UP000002320 UP000075840 UP000075881 UP000075903 UP000007062 UP000069272 UP000075886 UP000075920 UP000075880 UP000183832 UP000075883 UP000075900 UP000000673 UP000053105 UP000192223 UP000007819 UP000076408 UP000078542 UP000075884 UP000242457 UP000037069 UP000000311 UP000005203 UP000007755 UP000078540 UP000005205 UP000008237 UP000053097 UP000279307 UP000027135 UP000091820 UP000078492 UP000078541 UP000009046 UP000095300 UP000092444 UP000075809 UP000009192 UP000192221 UP000095301 UP000268350 UP000001292 UP000008711 UP000008792 UP000000803 UP000001819
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JIQ1
A0A0N1IBZ5
A0A0N0PAJ1
A0A2A4JZQ1
A0A212FC21
A0A1E1WKK5
+ More
A0A1E1WUZ1 A0A2A4K0U0 A0A1S4FAM4 A0A1S4FAU2 A0A1L8DPS8 A0A1L8DPH4 Q17A21 Q17A20 A0A182G1X8 A0A0L7KX63 K7J8G5 A0A1Q3FIW5 A0A1Q3FJ17 A0A1Q3FJ00 A0A1Q3FJ26 A0A084VQK1 A0A336M1P2 A0A182P7W7 B0XCS0 A0A182IA85 A0A182JSU7 A0A182UY80 Q7QFL4 A0A182FIZ9 A0A182QHW0 A0A1S4G989 A0A182W4P0 A0A182JG23 A0A1J1HSW6 A0A182MJE9 A0A182RBH7 W5JLJ6 A0A1J1HSV2 A0A336M555 A0A0M9A432 A0A1W4X666 A0A1W4XFP3 A0A1W4XGL2 A0A2H8TDI6 J9JV68 A0A2S2QM76 A0A182Y7J4 A0A0C9QJR1 A0A151I9C3 A0A182N115 A0A1B6DF11 A0A1B6ECQ7 A0A2A3ETZ4 A0A0L0BY61 E2AP22 A0A088AMW9 F4X0V1 A0A151HZ41 A0A158N9E9 E2BB29 A0A1Y1KJZ8 A0A026WGR5 A0A067RID9 A0A1A9WZ27 T1PGP1 A0A195E220 A0A151JV66 A0A1L8EAD8 E0VDP6 A0A1I8P1Y8 A0A224XA01 A0A1I8P260 A0A1L8EHL7 A0A1B0FP21 T1PEZ0 A0A151X2L5 A0A1Y1KK87 T1PEU8 T1PHE4 A0A1L8EAN0 A0A034UZJ8 A0A1L8EA36 B4K6G9 A0A1W4U686 A0A1I8M7B4 A0A1W4UHW4 A0A1W4UHZ5 A0A3B0JKH5 B4HZZ6 A0A3B0JEL6 A0A0K8WM51 B3P7S3 B4M5V3 A0A3B0JJ83 Q0KHY3 Q0KHY3-1 I5AMT6
A0A1E1WUZ1 A0A2A4K0U0 A0A1S4FAM4 A0A1S4FAU2 A0A1L8DPS8 A0A1L8DPH4 Q17A21 Q17A20 A0A182G1X8 A0A0L7KX63 K7J8G5 A0A1Q3FIW5 A0A1Q3FJ17 A0A1Q3FJ00 A0A1Q3FJ26 A0A084VQK1 A0A336M1P2 A0A182P7W7 B0XCS0 A0A182IA85 A0A182JSU7 A0A182UY80 Q7QFL4 A0A182FIZ9 A0A182QHW0 A0A1S4G989 A0A182W4P0 A0A182JG23 A0A1J1HSW6 A0A182MJE9 A0A182RBH7 W5JLJ6 A0A1J1HSV2 A0A336M555 A0A0M9A432 A0A1W4X666 A0A1W4XFP3 A0A1W4XGL2 A0A2H8TDI6 J9JV68 A0A2S2QM76 A0A182Y7J4 A0A0C9QJR1 A0A151I9C3 A0A182N115 A0A1B6DF11 A0A1B6ECQ7 A0A2A3ETZ4 A0A0L0BY61 E2AP22 A0A088AMW9 F4X0V1 A0A151HZ41 A0A158N9E9 E2BB29 A0A1Y1KJZ8 A0A026WGR5 A0A067RID9 A0A1A9WZ27 T1PGP1 A0A195E220 A0A151JV66 A0A1L8EAD8 E0VDP6 A0A1I8P1Y8 A0A224XA01 A0A1I8P260 A0A1L8EHL7 A0A1B0FP21 T1PEZ0 A0A151X2L5 A0A1Y1KK87 T1PEU8 T1PHE4 A0A1L8EAN0 A0A034UZJ8 A0A1L8EA36 B4K6G9 A0A1W4U686 A0A1I8M7B4 A0A1W4UHW4 A0A1W4UHZ5 A0A3B0JKH5 B4HZZ6 A0A3B0JEL6 A0A0K8WM51 B3P7S3 B4M5V3 A0A3B0JJ83 Q0KHY3 Q0KHY3-1 I5AMT6
Ontologies
GO
Topology
Subcellular location
Membrane
Cell junction
Septate junction
Lateral cell membrane
Apicolateral cell membrane
Cell junction
Septate junction
Lateral cell membrane
Apicolateral cell membrane
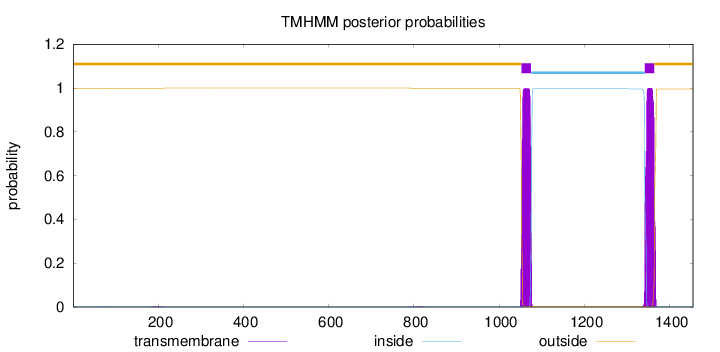
Length:
1455
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
45.88227
Exp number, first 60 AAs:
0.0015
Total prob of N-in:
0.00350
outside
1 - 1052
TMhelix
1053 - 1075
inside
1076 - 1341
TMhelix
1342 - 1364
outside
1365 - 1455
Population Genetic Test Statistics
Pi
251.634775
Theta
191.91587
Tajima's D
1.037677
CLR
0.342809
CSRT
0.669216539173041
Interpretation
Uncertain
