Gene
KWMTBOMO08444
Pre Gene Modal
BGIBMGA009403
Annotation
Glycosyltransferase-like_protein_LARGE2_[Papilio_xuthus]
Full name
LARGE xylosyl- and glucuronyltransferase 1
+ More
LARGE xylosyl- and glucuronyltransferase 2
LARGE xylosyl- and glucuronyltransferase 2
Alternative Name
Acetylglucosaminyltransferase-like 1A
Glycosyltransferase-like protein
Glycosyltransferase-like 1B
Glycosyltransferase-like protein
Glycosyltransferase-like 1B
Location in the cell
Extracellular Reliability : 1.11 PlasmaMembrane Reliability : 1.412
Sequence
CDS
ATGAAAGAAATTTCAGTAGAATATACTACCACATATCAAGTTCCTGTTATACCAGATTCTATAGGCACCTTGTGTGAAACAATTCATATAGCATTAGTTTGTATGGGTAAATGTACGAGACTGATCACTCCTATGTTTAAATCGTTGTTACACAACAGACAGAATCCGTTACATTTTCACATGGTCGTGGATAATACTGCAGAGCACACAGTCGCTGTGCTATTTGATACTTGGATGCTACCTGACGTAAAATACACATGCTACAACGCGCAGAATCGATTGTCCGAAGTCAGATGGATTCCCAACAGTCATTATTCCGGTGTATACGCTCTGGTCAAGCTGTTGTTCCCGGATATCCTGCCAGAGAACCTGGACCGGATCATTGTGTTGGACTCGGATTTGACCTTTCTCAGTGACGTAGCTGAGATCTGGAGGATGTTCAGGAACATGACCAACGAACAGTTTATAGGTTTGGTGGAAAACGAAAGCAATTGGTATTACGACACGGCGAAGCGATGGCCCGCGCTGGGCAGGGGCTTCAACACCGGCCTGATGCTGCTGGACTTGAAAAAGATCAGAACGAACATAGACTGGACTGATCTATGGCAGAACGCCATCAAGGAGAACATAGAGTCGCTGAAGCAGACGACCCTCGCCGACCAGGACATTATTAATGCCATCATAAAGAAACATCCTAATTACGTTTATAAAATTGGGTGTCAGTACAATGTGCAAATGTCGACGAAGACACTGTCCAAAAATTGTTATCGTGAAAATAAAAATAATGTTAAGGTCGTTCACTGGAATTCTCCAGCGAAGTACAATATAAAGATCAGAGACGCGCACTATTTCCGTAATCTATATCAGAGCTATGTTAATTTTGACGGGAATCTGTTGAGGGAGAAACTACATACGTGCAGTCAGAATGAAGCGCTGACTAATAAGATAAATTACTCCGATCTGTGTGTTAGTTTTCGTTCGGCTCAGAACATTAAGCTGCGTACTCACTTGTACTACATGGACTACAGCTACAATAATATCGACGACTTCGACGTCACCTTAGTTCTGCAGCTGTCCATGGACCGACTGCAGTTCCTGGAGAGACTTGTCAATCATTGGGAAGGTCCGTTAAGTGCAGCGATATATTTGTCGGACTGTGACGTCACGAAGCTGGAGAGCTTCACGCGGGACTGGTCGGACACGCTAAGCTCCCGGAAGAACATCGGATACCACTTAGTATTCAAACATGATTCGGTACATTACCCGGTGAATTATCTCCGTAACGTCGCGCTGGAGAATGTGAACACGCCCTACGTGTTCCTCATGGACGTCGACTTCGTTCCCATGGCGGGCCTGTACCAACATCTCAGGGACGCGATACGACTCATAAACCCTTATCCACAGAAAAAGTGTCTGGTGGTGCCCGCGTTCGAGACGCAGCGGTACCGCGCGTCCGTGCCGCGCGACAAGGCGGCGCTGCTGGCGCGGCTGGCGGCGCGGCACGGCGCCGGGGACGTCGCGCCCTTCCGCACCCGGGAGTGGCCGCGCGGACACCGGGCCACCAACTACACGAGGTGGATGACCGCCACTGCTCCTTATGAGGTGGAGTGGGAGTCCGACTACGAGCCCTACCTGGTGGTGCACCGGTCGGTACCTAAATACGACACCAGATTCTCGGGCTTCGGCTGGAACAAGGTGTCGCACAGCGTGGAGCTGCGCGCGACCGGGCACCGCGCGCTCGTGCTGCCGGCCGCGTTCCTCGTGCACACGCCGCACGCGCCCTCGCCCGACATCACCGCCTTCCGCGCCGACCCGCACTACCGCGTTTGTCTCGCGTTGTTGAAACAAGAATTCATGGACGACCTGAGCAGGAAGTATAACATGACCCTGGGAGGACCTTCGAAATTGGACAGTGTGTACCTCGGACAGGCCAAGAGCTTGCTGCTGAACCAGGCTAAGGAGTTGCCTTAG
Protein
MKEISVEYTTTYQVPVIPDSIGTLCETIHIALVCMGKCTRLITPMFKSLLHNRQNPLHFHMVVDNTAEHTVAVLFDTWMLPDVKYTCYNAQNRLSEVRWIPNSHYSGVYALVKLLFPDILPENLDRIIVLDSDLTFLSDVAEIWRMFRNMTNEQFIGLVENESNWYYDTAKRWPALGRGFNTGLMLLDLKKIRTNIDWTDLWQNAIKENIESLKQTTLADQDIINAIIKKHPNYVYKIGCQYNVQMSTKTLSKNCYRENKNNVKVVHWNSPAKYNIKIRDAHYFRNLYQSYVNFDGNLLREKLHTCSQNEALTNKINYSDLCVSFRSAQNIKLRTHLYYMDYSYNNIDDFDVTLVLQLSMDRLQFLERLVNHWEGPLSAAIYLSDCDVTKLESFTRDWSDTLSSRKNIGYHLVFKHDSVHYPVNYLRNVALENVNTPYVFLMDVDFVPMAGLYQHLRDAIRLINPYPQKKCLVVPAFETQRYRASVPRDKAALLARLAARHGAGDVAPFRTREWPRGHRATNYTRWMTATAPYEVEWESDYEPYLVVHRSVPKYDTRFSGFGWNKVSHSVELRATGHRALVLPAAFLVHTPHAPSPDITAFRADPHYRVCLALLKQEFMDDLSRKYNMTLGGPSKLDSVYLGQAKSLLLNQAKELP
Summary
Description
Bifunctional glycosyltransferase with both xylosyltransferase and beta-1,3-glucuronyltransferase activities involved in the biosynthesis of the phosphorylated O-mannosyl trisaccharide (N-acetylgalactosamine-beta-3-N-acetylglucosamine-beta-4-(phosphate-6-)mannose), a carbohydrate structure present in alpha-dystroglycan (DAG1) (PubMed:23125099, PubMed:23135544). Phosphorylated O-mannosyl trisaccharid is required for binding laminin G-like domain-containing extracellular proteins with high affinity and plays a key role in skeletal muscle function and regeneration (PubMed:15184894, PubMed:24132234). LARGE elongates the glucuronyl-beta-1,4-xylose-beta disaccharide primer structure initiated by B3GNT1/B4GAT1 by adding repeating units [-3-Xylose-alpha-1,3-GlcA-beta-1-] to produce a heteropolysaccharide (By similarity).
Bifunctional glycosyltransferase with both xylosyltransferase and beta-1,3-glucuronyltransferase activities involved in the biosynthesis of the phosphorylated O-mannosyl trisaccharide (N-acetylgalactosamine-beta-3-N-acetylglucosamine-beta-4-(phosphate-6-)mannose), a carbohydrate structure present in alpha-dystroglycan (DAG1). Phosphorylated O-mannosyl trisaccharid is required for binding laminin G-like domain-containing extracellular proteins with high affinity. Elongates the glucuronyl-beta-1,4-xylose-beta disaccharide primer structure by adding repeating units [-3-Xylose-alpha-1,3-GlcA-beta-1-] to produce a heteropolysaccharide. Has a higher activity toward alpha-dystroglycan than LARGE.
Bifunctional glycosyltransferase with both xylosyltransferase and beta-1,3-glucuronyltransferase activities involved in the biosynthesis of the phosphorylated O-mannosyl trisaccharide (N-acetylgalactosamine-beta-3-N-acetylglucosamine-beta-4-(phosphate-6-)mannose), a carbohydrate structure present in alpha-dystroglycan (DAG1) (PubMed:22223806). Phosphorylated O-mannosyl trisaccharid is required for binding laminin G-like domain-containing extracellular proteins with high affinity and plays a key role in skeletal muscle function and regeneration. LARGE elongates the glucuronyl-beta-1,4-xylose-beta disaccharide primer structure initiated by B3GNT1/B4GAT1 by adding repeating units [-3-Xylose-alpha-1,3-GlcA-beta-1-] to produce a heteropolysaccharide (PubMed:25279699).
Bifunctional glycosyltransferase with both xylosyltransferase and beta-1,3-glucuronyltransferase activities involved in the biosynthesis of the phosphorylated O-mannosyl trisaccharide (N-acetylgalactosamine-beta-3-N-acetylglucosamine-beta-4-(phosphate-6-)mannose), a carbohydrate structure present in alpha-dystroglycan (DAG1). Phosphorylated O-mannosyl trisaccharid is required for binding laminin G-like domain-containing extracellular proteins with high affinity. Elongates the glucuronyl-beta-1,4-xylose-beta disaccharide primer structure by adding repeating units [-3-Xylose-alpha-1,3-GlcA-beta-1-] to produce a heteropolysaccharide. Has a higher activity toward alpha-dystroglycan than LARGE.
Bifunctional glycosyltransferase with both xylosyltransferase and beta-1,3-glucuronyltransferase activities involved in the biosynthesis of the phosphorylated O-mannosyl trisaccharide (N-acetylgalactosamine-beta-3-N-acetylglucosamine-beta-4-(phosphate-6-)mannose), a carbohydrate structure present in alpha-dystroglycan (DAG1) (PubMed:22223806). Phosphorylated O-mannosyl trisaccharid is required for binding laminin G-like domain-containing extracellular proteins with high affinity and plays a key role in skeletal muscle function and regeneration. LARGE elongates the glucuronyl-beta-1,4-xylose-beta disaccharide primer structure initiated by B3GNT1/B4GAT1 by adding repeating units [-3-Xylose-alpha-1,3-GlcA-beta-1-] to produce a heteropolysaccharide (PubMed:25279699).
Cofactor
Mn(2+)
Subunit
Interacts with DAG1 (via the N-terminal domain of alpha-DAG1); the interaction increases binding of DAG1 to laminin. Interacts with B3GNT1/B4GAT1.
Interacts with DAG1 (via the N-terminal domain of alpha-DAG1); the interaction increases binding of DAG1 to laminin (By similarity). Interacts with B3GNT1/B4GAT1 (PubMed:19587235).
Interacts with DAG1 (via the N-terminal domain of alpha-DAG1); the interaction increases binding of DAG1 to laminin (By similarity). Interacts with B3GNT1/B4GAT1 (PubMed:19587235).
Similarity
In the C-terminal section; belongs to the glycosyltransferase 49 family.
In the N-terminal section; belongs to the glycosyltransferase 8 family.
In the N-terminal section; belongs to the glycosyltransferase 8 family.
Keywords
Coiled coil
Complete proteome
Glycoprotein
Glycosyltransferase
Golgi apparatus
Manganese
Membrane
Metal-binding
Multifunctional enzyme
Reference proteome
Signal-anchor
Transferase
Transmembrane
Transmembrane helix
Alternative splicing
Congenital muscular dystrophy
Disease mutation
Dystroglycanopathy
Lissencephaly
Polymorphism
Feature
chain LARGE xylosyl- and glucuronyltransferase 1
splice variant In isoform 2.
sequence variant In dbSNP:rs470035.
splice variant In isoform 2.
sequence variant In dbSNP:rs470035.
Uniprot
A0A437BH37
A0A0N1PHL8
A0A2A4K034
A0A212FC44
A0A2H1VJT8
A0A2H1V8R8
+ More
A0A0N0PE39 A0A2L2Y9Z4 T1JEW5 E0VGD3 R7TJK9 A0A1B6CYE9 A0A1D2N9G8 A0A0A9YJR9 A0A3S3QIJ1 A0A0G2K618 A0A023F5B6 K7FUI6 Q059X9 Q9Z1M7 F7BXD5 A0A147BEK3 H0WH59 B3DFN1 A0A1S3FWF7 A0A250Y4X4 A0A1U7U2V0 A0A2K6EKN8 A0A3S1BCW5 Q66PG1 G1SG12 A0A2K5YPC0 A0A2Y9GVR2 A0A2J7PG24 A0A1U8DIB7 A0A3Q0GZD9 A0A1A7YWM9 G3TKC6 A0A2J7PG28 V5IHD6 I3MWQ4 G1MCE4 F1P723 A0A2K5C7A4 M3WNA3 A0A3Q7TN28 M3YLT6 A0A2K5PFQ5 G7N3P5 A0A2J8UVN3 A0A2K6TC39 A0A3P8XNA6 H3AH90 A0A1S3KT15 A0A0D9R7A4 A0A2K6PR53 A0A2Y9KZW9 A0A3Q7V2H6 G7PFA2 A0A2K5P157 A0A2K6AN31 A0A2K6K897 A0A096NJQ6 F6QJZ2 H2QZS2 A0A1Z5KUC0 X5DR28 O95461 A0A151NHX1 A0A452ISM3 A0A3Q3JPB6 H0VRT1 G1P039 L5LIU7 A0A402EM90 F6W6X6 A0A2U3X4A5 A0A067QRU2 A0A1S3PRW3 K9INM3 T1HYF4 A0A060WGJ6 A0A1A8MAF7 A0A1A7XMX1 A0A3B4CR51 A0A3B3XA23 A0A452ERG5 A0A3Q2I5B9 E1BDI0 A0A1A8BHC4 A0A091IPA3 A0A1A8KBF5 A0A1A8B0W5 A0A1A8S7R4 A0A2D0QX56 A0A226E869 M3ZES1 A0A1V4K7K2 A0A0F7Z8J2 A0A3Q2PUR7
A0A0N0PE39 A0A2L2Y9Z4 T1JEW5 E0VGD3 R7TJK9 A0A1B6CYE9 A0A1D2N9G8 A0A0A9YJR9 A0A3S3QIJ1 A0A0G2K618 A0A023F5B6 K7FUI6 Q059X9 Q9Z1M7 F7BXD5 A0A147BEK3 H0WH59 B3DFN1 A0A1S3FWF7 A0A250Y4X4 A0A1U7U2V0 A0A2K6EKN8 A0A3S1BCW5 Q66PG1 G1SG12 A0A2K5YPC0 A0A2Y9GVR2 A0A2J7PG24 A0A1U8DIB7 A0A3Q0GZD9 A0A1A7YWM9 G3TKC6 A0A2J7PG28 V5IHD6 I3MWQ4 G1MCE4 F1P723 A0A2K5C7A4 M3WNA3 A0A3Q7TN28 M3YLT6 A0A2K5PFQ5 G7N3P5 A0A2J8UVN3 A0A2K6TC39 A0A3P8XNA6 H3AH90 A0A1S3KT15 A0A0D9R7A4 A0A2K6PR53 A0A2Y9KZW9 A0A3Q7V2H6 G7PFA2 A0A2K5P157 A0A2K6AN31 A0A2K6K897 A0A096NJQ6 F6QJZ2 H2QZS2 A0A1Z5KUC0 X5DR28 O95461 A0A151NHX1 A0A452ISM3 A0A3Q3JPB6 H0VRT1 G1P039 L5LIU7 A0A402EM90 F6W6X6 A0A2U3X4A5 A0A067QRU2 A0A1S3PRW3 K9INM3 T1HYF4 A0A060WGJ6 A0A1A8MAF7 A0A1A7XMX1 A0A3B4CR51 A0A3B3XA23 A0A452ERG5 A0A3Q2I5B9 E1BDI0 A0A1A8BHC4 A0A091IPA3 A0A1A8KBF5 A0A1A8B0W5 A0A1A8S7R4 A0A2D0QX56 A0A226E869 M3ZES1 A0A1V4K7K2 A0A0F7Z8J2 A0A3Q2PUR7
EC Number
2.4.-.-
Pubmed
26354079
22118469
26561354
20566863
23254933
27289101
+ More
25401762 26823975 15057822 25474469 17381049 12040188 15489334 9892679 12693553 11381262 15210115 15958417 15184894 23125099 23135544 24132234 25138275 25243066 29652888 28087693 23594743 21993624 25765539 20010809 16341006 17975172 22002653 25069045 9215903 25362486 17431167 25319552 15164055 16136131 28528879 11181995 24722188 9628581 15461802 10591208 15752776 15661757 19587235 21987822 22223806 25279699 25279697 12966029 19067344 19299310 24709677 22293439 28562605 19892987 24845553 24755649 19393038 23542700
25401762 26823975 15057822 25474469 17381049 12040188 15489334 9892679 12693553 11381262 15210115 15958417 15184894 23125099 23135544 24132234 25138275 25243066 29652888 28087693 23594743 21993624 25765539 20010809 16341006 17975172 22002653 25069045 9215903 25362486 17431167 25319552 15164055 16136131 28528879 11181995 24722188 9628581 15461802 10591208 15752776 15661757 19587235 21987822 22223806 25279699 25279697 12966029 19067344 19299310 24709677 22293439 28562605 19892987 24845553 24755649 19393038 23542700
EMBL
RSAL01000059
RVE49710.1
KQ458751
KPJ05299.1
NWSH01000337
PCG77288.1
+ More
AGBW02009237 OWR51287.1 ODYU01002686 SOQ40514.1 ODYU01001253 SOQ37207.1 KQ459986 KPJ18829.1 IAAA01016561 IAAA01016562 LAA04992.1 JH432130 DS235137 EEB12439.1 AMQN01013876 KB310564 ELT91285.1 GEDC01018877 JAS18421.1 LJIJ01000134 ODN01913.1 GBHO01011738 GDHC01020207 JAG31866.1 JAP98421.1 NCKU01002576 RWS09293.1 AABR07042861 AABR07042862 AABR07042863 AABR07042864 AABR07042865 AABR07042866 GBBI01002408 JAC16304.1 AGCU01195138 AGCU01195139 AGCU01195140 AGCU01195141 AGCU01195142 AGCU01195143 AGCU01195144 AGCU01195145 AGCU01195146 AGCU01195147 BC125487 BC125489 CH466525 AAI25490.1 EDL10819.1 AJ006278 AK122328 BC061506 BC100399 GAMT01006968 GAMS01004399 GAMR01008255 GAMQ01004062 GAMQ01004061 GAMP01003926 GAMP01003925 GAMP01003924 JAB04893.1 JAB18737.1 JAB25677.1 JAB37789.1 JAB48829.1 GEGO01006659 JAR88745.1 AAQR03016478 AAQR03016479 AAQR03016480 AAQR03016481 AAQR03016482 BC162103 AAI62103.1 GFFV01001295 JAV38650.1 RQTK01000359 RUS81043.1 AY662339 BX908385 BX936304 AAGW02060986 AAGW02060987 AAGW02060988 AAGW02060989 AAGW02060990 AAGW02060991 AAGW02060992 NEVH01025635 PNF15282.1 HADX01012128 SBP34360.1 PNF15281.1 GANP01005122 JAB79346.1 AGTP01024453 AGTP01024454 AGTP01024455 AGTP01024456 AGTP01024457 AGTP01024458 AGTP01024459 AGTP01024460 AGTP01024461 AGTP01024462 ACTA01075690 ACTA01083690 ACTA01091690 ACTA01099690 ACTA01107690 ACTA01115690 ACTA01123690 ACTA01131690 ACTA01139690 ACTA01147689 AAEX03007366 AANG04003655 AEYP01012497 AEYP01012498 AEYP01012499 AEYP01012500 AEYP01012501 AEYP01012502 AEYP01012503 AEYP01012504 AEYP01012505 AEYP01012506 CM001262 EHH20177.1 ABGA01343582 ABGA01343583 ABGA01343584 ABGA01343585 ABGA01343586 ABGA01343587 ABGA01343588 ABGA01343589 ABGA01343590 ABGA01343591 NDHI03003442 PNJ49333.1 PNJ49334.1 AFYH01014743 AFYH01014744 AFYH01014745 AFYH01014746 AFYH01014747 AFYH01014748 AFYH01014749 AFYH01014750 AFYH01014751 AFYH01014752 AQIB01136734 AQIB01136735 AQIB01136736 AQIB01136737 AQIB01136738 AQIB01136739 AQIB01136740 AQIB01136741 AQIA01007180 AQIA01007181 AQIA01007182 AQIA01007183 AQIA01007184 AQIA01007185 AQIA01007186 AQIA01007187 AQIA01007188 CM001285 EHH65786.1 AHZZ02001146 AHZZ02001147 AHZZ02001148 AHZZ02001149 AHZZ02001150 AHZZ02001151 AHZZ02001152 AHZZ02001153 JSUE03005380 JSUE03005381 JSUE03005382 JSUE03005383 JSUE03005384 JSUE03005385 JSUE03005386 JSUE03005387 JSUE03005388 JSUE03005389 JU337091 AFE80844.1 AACZ04068072 GABC01005918 GABC01005917 GABF01000316 GABF01000315 GABD01008726 GABD01008725 GABE01009849 GABE01009848 NBAG03000026 JAA05420.1 JAA21829.1 JAA24374.1 JAA34890.1 PNI98083.1 PNI98084.1 GFJQ02008355 JAV98614.1 KJ534877 CH471095 AHW56517.1 EAW60041.1 AJ007583 AB011181 CR456510 AL008630 AL008715 AL096754 Z68287 Z69042 Z69943 Z70288 Z82173 BC117425 BC126404 AKHW03002956 KYO36407.1 AAKN02054133 AAPE02024977 AAPE02024978 AAPE02024979 AAPE02024980 AAPE02024981 AAPE02024982 AAPE02024983 AAPE02024984 AAPE02024985 AAPE02024986 KB111329 ELK25930.1 BDOT01000023 GCF45258.1 KK853033 KDR12249.1 GABZ01004573 JAA48952.1 ACPB03007941 FR904535 CDQ66232.1 HAEF01012155 SBR53314.1 HADW01018032 SBP19432.1 LWLT01000005 HADZ01002352 SBP66293.1 KK500774 KFP10182.1 HAEE01009582 SBR29632.1 HADY01022083 SBP60568.1 HAEI01011532 SBS13594.1 LNIX01000006 OXA53187.1 LSYS01004200 OPJ80414.1 GBEX01001759 JAI12801.1
AGBW02009237 OWR51287.1 ODYU01002686 SOQ40514.1 ODYU01001253 SOQ37207.1 KQ459986 KPJ18829.1 IAAA01016561 IAAA01016562 LAA04992.1 JH432130 DS235137 EEB12439.1 AMQN01013876 KB310564 ELT91285.1 GEDC01018877 JAS18421.1 LJIJ01000134 ODN01913.1 GBHO01011738 GDHC01020207 JAG31866.1 JAP98421.1 NCKU01002576 RWS09293.1 AABR07042861 AABR07042862 AABR07042863 AABR07042864 AABR07042865 AABR07042866 GBBI01002408 JAC16304.1 AGCU01195138 AGCU01195139 AGCU01195140 AGCU01195141 AGCU01195142 AGCU01195143 AGCU01195144 AGCU01195145 AGCU01195146 AGCU01195147 BC125487 BC125489 CH466525 AAI25490.1 EDL10819.1 AJ006278 AK122328 BC061506 BC100399 GAMT01006968 GAMS01004399 GAMR01008255 GAMQ01004062 GAMQ01004061 GAMP01003926 GAMP01003925 GAMP01003924 JAB04893.1 JAB18737.1 JAB25677.1 JAB37789.1 JAB48829.1 GEGO01006659 JAR88745.1 AAQR03016478 AAQR03016479 AAQR03016480 AAQR03016481 AAQR03016482 BC162103 AAI62103.1 GFFV01001295 JAV38650.1 RQTK01000359 RUS81043.1 AY662339 BX908385 BX936304 AAGW02060986 AAGW02060987 AAGW02060988 AAGW02060989 AAGW02060990 AAGW02060991 AAGW02060992 NEVH01025635 PNF15282.1 HADX01012128 SBP34360.1 PNF15281.1 GANP01005122 JAB79346.1 AGTP01024453 AGTP01024454 AGTP01024455 AGTP01024456 AGTP01024457 AGTP01024458 AGTP01024459 AGTP01024460 AGTP01024461 AGTP01024462 ACTA01075690 ACTA01083690 ACTA01091690 ACTA01099690 ACTA01107690 ACTA01115690 ACTA01123690 ACTA01131690 ACTA01139690 ACTA01147689 AAEX03007366 AANG04003655 AEYP01012497 AEYP01012498 AEYP01012499 AEYP01012500 AEYP01012501 AEYP01012502 AEYP01012503 AEYP01012504 AEYP01012505 AEYP01012506 CM001262 EHH20177.1 ABGA01343582 ABGA01343583 ABGA01343584 ABGA01343585 ABGA01343586 ABGA01343587 ABGA01343588 ABGA01343589 ABGA01343590 ABGA01343591 NDHI03003442 PNJ49333.1 PNJ49334.1 AFYH01014743 AFYH01014744 AFYH01014745 AFYH01014746 AFYH01014747 AFYH01014748 AFYH01014749 AFYH01014750 AFYH01014751 AFYH01014752 AQIB01136734 AQIB01136735 AQIB01136736 AQIB01136737 AQIB01136738 AQIB01136739 AQIB01136740 AQIB01136741 AQIA01007180 AQIA01007181 AQIA01007182 AQIA01007183 AQIA01007184 AQIA01007185 AQIA01007186 AQIA01007187 AQIA01007188 CM001285 EHH65786.1 AHZZ02001146 AHZZ02001147 AHZZ02001148 AHZZ02001149 AHZZ02001150 AHZZ02001151 AHZZ02001152 AHZZ02001153 JSUE03005380 JSUE03005381 JSUE03005382 JSUE03005383 JSUE03005384 JSUE03005385 JSUE03005386 JSUE03005387 JSUE03005388 JSUE03005389 JU337091 AFE80844.1 AACZ04068072 GABC01005918 GABC01005917 GABF01000316 GABF01000315 GABD01008726 GABD01008725 GABE01009849 GABE01009848 NBAG03000026 JAA05420.1 JAA21829.1 JAA24374.1 JAA34890.1 PNI98083.1 PNI98084.1 GFJQ02008355 JAV98614.1 KJ534877 CH471095 AHW56517.1 EAW60041.1 AJ007583 AB011181 CR456510 AL008630 AL008715 AL096754 Z68287 Z69042 Z69943 Z70288 Z82173 BC117425 BC126404 AKHW03002956 KYO36407.1 AAKN02054133 AAPE02024977 AAPE02024978 AAPE02024979 AAPE02024980 AAPE02024981 AAPE02024982 AAPE02024983 AAPE02024984 AAPE02024985 AAPE02024986 KB111329 ELK25930.1 BDOT01000023 GCF45258.1 KK853033 KDR12249.1 GABZ01004573 JAA48952.1 ACPB03007941 FR904535 CDQ66232.1 HAEF01012155 SBR53314.1 HADW01018032 SBP19432.1 LWLT01000005 HADZ01002352 SBP66293.1 KK500774 KFP10182.1 HAEE01009582 SBR29632.1 HADY01022083 SBP60568.1 HAEI01011532 SBS13594.1 LNIX01000006 OXA53187.1 LSYS01004200 OPJ80414.1 GBEX01001759 JAI12801.1
Proteomes
UP000283053
UP000053268
UP000218220
UP000007151
UP000053240
UP000009046
+ More
UP000014760 UP000094527 UP000285301 UP000002494 UP000007267 UP000000589 UP000008225 UP000005225 UP000081671 UP000189704 UP000233160 UP000271974 UP000000437 UP000001811 UP000233140 UP000248481 UP000235965 UP000189705 UP000007646 UP000005215 UP000008912 UP000002254 UP000233020 UP000011712 UP000286640 UP000000715 UP000233040 UP000001595 UP000233220 UP000265140 UP000008672 UP000087266 UP000029965 UP000233200 UP000248482 UP000286642 UP000009130 UP000233100 UP000233060 UP000233120 UP000233180 UP000028761 UP000006718 UP000002277 UP000005640 UP000050525 UP000291020 UP000261600 UP000005447 UP000001074 UP000288954 UP000002281 UP000245340 UP000027135 UP000015103 UP000193380 UP000261440 UP000261480 UP000291000 UP000009136 UP000053119 UP000221080 UP000198287 UP000002852 UP000190648 UP000265000
UP000014760 UP000094527 UP000285301 UP000002494 UP000007267 UP000000589 UP000008225 UP000005225 UP000081671 UP000189704 UP000233160 UP000271974 UP000000437 UP000001811 UP000233140 UP000248481 UP000235965 UP000189705 UP000007646 UP000005215 UP000008912 UP000002254 UP000233020 UP000011712 UP000286640 UP000000715 UP000233040 UP000001595 UP000233220 UP000265140 UP000008672 UP000087266 UP000029965 UP000233200 UP000248482 UP000286642 UP000009130 UP000233100 UP000233060 UP000233120 UP000233180 UP000028761 UP000006718 UP000002277 UP000005640 UP000050525 UP000291020 UP000261600 UP000005447 UP000001074 UP000288954 UP000002281 UP000245340 UP000027135 UP000015103 UP000193380 UP000261440 UP000261480 UP000291000 UP000009136 UP000053119 UP000221080 UP000198287 UP000002852 UP000190648 UP000265000
Pfam
PF01501 Glyco_transf_8
SUPFAM
SSF53448
SSF53448
Gene 3D
ProteinModelPortal
A0A437BH37
A0A0N1PHL8
A0A2A4K034
A0A212FC44
A0A2H1VJT8
A0A2H1V8R8
+ More
A0A0N0PE39 A0A2L2Y9Z4 T1JEW5 E0VGD3 R7TJK9 A0A1B6CYE9 A0A1D2N9G8 A0A0A9YJR9 A0A3S3QIJ1 A0A0G2K618 A0A023F5B6 K7FUI6 Q059X9 Q9Z1M7 F7BXD5 A0A147BEK3 H0WH59 B3DFN1 A0A1S3FWF7 A0A250Y4X4 A0A1U7U2V0 A0A2K6EKN8 A0A3S1BCW5 Q66PG1 G1SG12 A0A2K5YPC0 A0A2Y9GVR2 A0A2J7PG24 A0A1U8DIB7 A0A3Q0GZD9 A0A1A7YWM9 G3TKC6 A0A2J7PG28 V5IHD6 I3MWQ4 G1MCE4 F1P723 A0A2K5C7A4 M3WNA3 A0A3Q7TN28 M3YLT6 A0A2K5PFQ5 G7N3P5 A0A2J8UVN3 A0A2K6TC39 A0A3P8XNA6 H3AH90 A0A1S3KT15 A0A0D9R7A4 A0A2K6PR53 A0A2Y9KZW9 A0A3Q7V2H6 G7PFA2 A0A2K5P157 A0A2K6AN31 A0A2K6K897 A0A096NJQ6 F6QJZ2 H2QZS2 A0A1Z5KUC0 X5DR28 O95461 A0A151NHX1 A0A452ISM3 A0A3Q3JPB6 H0VRT1 G1P039 L5LIU7 A0A402EM90 F6W6X6 A0A2U3X4A5 A0A067QRU2 A0A1S3PRW3 K9INM3 T1HYF4 A0A060WGJ6 A0A1A8MAF7 A0A1A7XMX1 A0A3B4CR51 A0A3B3XA23 A0A452ERG5 A0A3Q2I5B9 E1BDI0 A0A1A8BHC4 A0A091IPA3 A0A1A8KBF5 A0A1A8B0W5 A0A1A8S7R4 A0A2D0QX56 A0A226E869 M3ZES1 A0A1V4K7K2 A0A0F7Z8J2 A0A3Q2PUR7
A0A0N0PE39 A0A2L2Y9Z4 T1JEW5 E0VGD3 R7TJK9 A0A1B6CYE9 A0A1D2N9G8 A0A0A9YJR9 A0A3S3QIJ1 A0A0G2K618 A0A023F5B6 K7FUI6 Q059X9 Q9Z1M7 F7BXD5 A0A147BEK3 H0WH59 B3DFN1 A0A1S3FWF7 A0A250Y4X4 A0A1U7U2V0 A0A2K6EKN8 A0A3S1BCW5 Q66PG1 G1SG12 A0A2K5YPC0 A0A2Y9GVR2 A0A2J7PG24 A0A1U8DIB7 A0A3Q0GZD9 A0A1A7YWM9 G3TKC6 A0A2J7PG28 V5IHD6 I3MWQ4 G1MCE4 F1P723 A0A2K5C7A4 M3WNA3 A0A3Q7TN28 M3YLT6 A0A2K5PFQ5 G7N3P5 A0A2J8UVN3 A0A2K6TC39 A0A3P8XNA6 H3AH90 A0A1S3KT15 A0A0D9R7A4 A0A2K6PR53 A0A2Y9KZW9 A0A3Q7V2H6 G7PFA2 A0A2K5P157 A0A2K6AN31 A0A2K6K897 A0A096NJQ6 F6QJZ2 H2QZS2 A0A1Z5KUC0 X5DR28 O95461 A0A151NHX1 A0A452ISM3 A0A3Q3JPB6 H0VRT1 G1P039 L5LIU7 A0A402EM90 F6W6X6 A0A2U3X4A5 A0A067QRU2 A0A1S3PRW3 K9INM3 T1HYF4 A0A060WGJ6 A0A1A8MAF7 A0A1A7XMX1 A0A3B4CR51 A0A3B3XA23 A0A452ERG5 A0A3Q2I5B9 E1BDI0 A0A1A8BHC4 A0A091IPA3 A0A1A8KBF5 A0A1A8B0W5 A0A1A8S7R4 A0A2D0QX56 A0A226E869 M3ZES1 A0A1V4K7K2 A0A0F7Z8J2 A0A3Q2PUR7
PDB
4WNH
E-value=2.62062e-07,
Score=133
Ontologies
PATHWAY
GO
GO:0016757
GO:0016021
GO:0016740
GO:0042285
GO:0030145
GO:0005794
GO:0035269
GO:0015020
GO:0043403
GO:0060538
GO:0046716
GO:0009101
GO:0043231
GO:0000139
GO:0006486
GO:0046872
GO:0008375
GO:0006044
GO:0030173
GO:0035252
GO:0006493
GO:0006688
GO:0005524
GO:0006468
GO:0003676
GO:0006182
GO:0051603
GO:0006412
GO:0016876
GO:0043039
Topology
Subcellular location
Golgi apparatus membrane
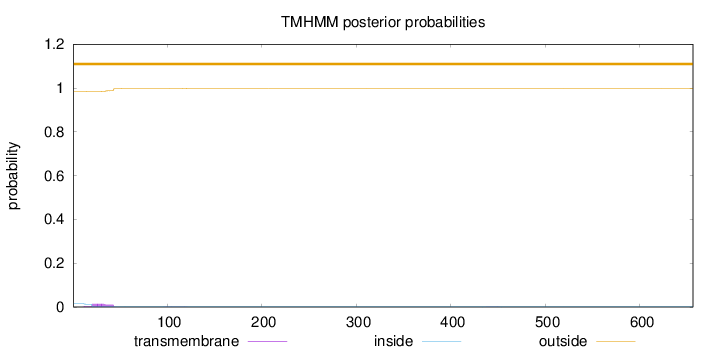
Length:
656
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.350820000000001
Exp number, first 60 AAs:
0.32967
Total prob of N-in:
0.01608
outside
1 - 656
Population Genetic Test Statistics
Pi
227.024865
Theta
188.91874
Tajima's D
0.940744
CLR
0.157828
CSRT
0.644517774111294
Interpretation
Uncertain
