Gene
KWMTBOMO08442
Annotation
parathyroid_hormone-responsive_B1_[Helicoverpa_armigera]
Location in the cell
Cytoplasmic Reliability : 1.726 Nuclear Reliability : 1.627
Sequence
CDS
ATGAAATGCTATTTCGGATGCCTTGATAACGGACTAAATTATGCAGCTCACATCGAAATTGATCAGCCACAGTTGTTGAGTGGAATTGTGCCGCCTTCAAGTATGAACAATATGATAAAACTTGAAAGTGTATCATACAGAATGAGGCTAATGCCTGACGAGAAAAGAATTGTGAATCTAATGTTTGATGAAATATCACTTTGTCCTGGAGTGACCTATGATCCTGATATAGATGCAGTGATAGGATTTGAGGATATGGGATCTAATAGTAGGAATAAAACTCTGGCTGACCATGCACTTGTATTTATGATTAAGAGTGTCAAAATTATTTGCGATCAAGCCAGGACAAATGTCAACGCACTAAAGAACTTACATCAGGATACAAGGGAGAAATATTTGAAGGAAGGACATGGAGATTACATAGAAAAAGCCATTGAAATTGGCGGAAAAAAAATATTTCCTCTATTTGATCCACCACATTTATTGAAAGGTGTGAGAAATAATTTACTGATCAAAGATGCTTTATTTGTGCAGGATGGTGTCATGAAGCGAGCAAAATGGGATCATATTAGAATGTTGTTGGATGTGGATGATGGTGATGACGAAATTAGATTAGTCAATAAACTGACGGAAGCACATGTTATTAAAGAAAAGATCCCAAAAATGAAAGTGAAATTTGCTGCACAAATATTCAGTCAAAGAGTCTCATCAGCTATGAGATTCCTTGCAAGTGAGTGTTGTATATGA
Protein
MKCYFGCLDNGLNYAAHIEIDQPQLLSGIVPPSSMNNMIKLESVSYRMRLMPDEKRIVNLMFDEISLCPGVTYDPDIDAVIGFEDMGSNSRNKTLADHALVFMIKSVKIICDQARTNVNALKNLHQDTREKYLKEGHGDYIEKAIEIGGKKIFPLFDPPHLLKGVRNNLLIKDALFVQDGVMKRAKWDHIRMLLDVDDGDDEIRLVNKLTEAHVIKEKIPKMKVKFAAQIFSQRVSSAMRFLASECCI
Summary
Uniprot
H9JIR1
F8SL48
A0A0N1IC55
A0A194QQL0
A0A2H1WVF6
A0A2H1WTP2
+ More
J9L1W2 X1WQI7 X1XTB8 J9M4P5 J9M961 J9LX62 J9L6Z3 J9KEP3 A0A2H1VJ77 A0A2A4JPP6 J9KEF2 J9M9N1 A0A3S2LLE6 J9LEP6 A0A2S2N697 J9M897 J9KSA4 X1XB84 J9LY75 A0A0T6BCB1 J9KBW7 X1XC48 J9LPQ0 J9KGM8 A0A2S2NC74 A0A1Y1NI49 A0A2S2P628 A0A2S2N6K3 A0A2H8TYS2 A0A0L7K5T4 A0A1Y1LY36 A0A2A4JYS8 A0A2A4JY71 A0A2A4JZ58 A0A0T6BAH8 X1XAX4 A0A3S2KX83 J9M6Y7 X1XMZ9 J9L3T7 A0A0L7LB47 A0A3S2LNK6 A0A2S2P5T9 J9L7T1 A0A437ASS2 J9KT02 X1XJT3 A0A3S2NSI9 J9KI00 A0A2S2P9C5 A0A3S2LM91 X1XGM9 A0A437BCW2 J9KFX0 A0A1Y1JZB1 A0A3S2NMS9 A0A139WA73 A0A139W9L5 A0A437BCX3 X1X0R9 J9L5B6 A0A3S2L9N1 J9KX16 D6X0C0 A0A0J7LAG3 A0A437BMU4 J9LQD8 A0A2S2QQP4 A0A1Y1KFJ5 A0A1Y1KI01 A0A1Y1KN14 A0A1Y1K6U6 A0A139W8P5 A0A3S2LEZ0 E9J9D7 E2AAV3 J9M7W9 A0A1Y1KLF8 N6U1G5 E9IPY0 A0A1Y1NHP2 A0A1Y1NHU8 A0A2J7RGL9 A0A2P8ZAL9
J9L1W2 X1WQI7 X1XTB8 J9M4P5 J9M961 J9LX62 J9L6Z3 J9KEP3 A0A2H1VJ77 A0A2A4JPP6 J9KEF2 J9M9N1 A0A3S2LLE6 J9LEP6 A0A2S2N697 J9M897 J9KSA4 X1XB84 J9LY75 A0A0T6BCB1 J9KBW7 X1XC48 J9LPQ0 J9KGM8 A0A2S2NC74 A0A1Y1NI49 A0A2S2P628 A0A2S2N6K3 A0A2H8TYS2 A0A0L7K5T4 A0A1Y1LY36 A0A2A4JYS8 A0A2A4JY71 A0A2A4JZ58 A0A0T6BAH8 X1XAX4 A0A3S2KX83 J9M6Y7 X1XMZ9 J9L3T7 A0A0L7LB47 A0A3S2LNK6 A0A2S2P5T9 J9L7T1 A0A437ASS2 J9KT02 X1XJT3 A0A3S2NSI9 J9KI00 A0A2S2P9C5 A0A3S2LM91 X1XGM9 A0A437BCW2 J9KFX0 A0A1Y1JZB1 A0A3S2NMS9 A0A139WA73 A0A139W9L5 A0A437BCX3 X1X0R9 J9L5B6 A0A3S2L9N1 J9KX16 D6X0C0 A0A0J7LAG3 A0A437BMU4 J9LQD8 A0A2S2QQP4 A0A1Y1KFJ5 A0A1Y1KI01 A0A1Y1KN14 A0A1Y1K6U6 A0A139W8P5 A0A3S2LEZ0 E9J9D7 E2AAV3 J9M7W9 A0A1Y1KLF8 N6U1G5 E9IPY0 A0A1Y1NHP2 A0A1Y1NHU8 A0A2J7RGL9 A0A2P8ZAL9
Pubmed
EMBL
BABH01032897
HQ645847
AEH16631.1
KQ461059
KPJ09927.1
KQ461175
+ More
KPJ07793.1 ODYU01011352 SOQ57041.1 ODYU01010582 SOQ55794.1 ABLF02008338 ABLF02016627 ABLF02063349 ABLF02008684 ABLF02008686 ABLF02008687 ABLF02011948 ABLF02019656 ABLF02019662 ABLF02041486 ABLF02009647 ABLF02009778 ABLF02009779 ABLF02025341 ABLF02042390 ABLF02017444 ODYU01002680 SOQ40482.1 NWSH01000959 PCG73380.1 ABLF02028679 ABLF02056730 ABLF02017692 ABLF02017693 RSAL01008431 RVE39798.1 ABLF02010326 ABLF02010327 GGMR01000046 MBY12665.1 ABLF02019592 ABLF02016553 ABLF02016555 ABLF02049531 ABLF02011758 ABLF02011759 ABLF02012658 LJIG01001987 KRT84953.1 ABLF02006763 ABLF02005638 ABLF02005639 ABLF02031840 ABLF02012220 ABLF02012222 ABLF02053218 GGMR01002126 MBY14745.1 GEZM01001796 JAV97521.1 GGMR01012308 MBY24927.1 GGMR01000178 MBY12797.1 GFXV01007611 MBW19416.1 JTDY01010331 KOB58244.1 GEZM01045715 JAV77678.1 NWSH01000363 PCG76969.1 PCG76967.1 PCG76968.1 LJIG01002596 KRT84344.1 ABLF02054892 RSAL01002057 RVE40596.1 ABLF02034357 ABLF02004413 ABLF02016452 ABLF02049812 ABLF02056148 ABLF02016989 JTDY01001960 KOB72446.1 RSAL01000885 RVE41073.1 GGMR01011939 MBY24558.1 ABLF02017696 RSAL01000699 RVE41218.1 ABLF02003651 ABLF02011697 RSAL01000257 RVE43416.1 ABLF02055473 ABLF02056006 GGMR01012897 MBY25516.1 RSAL01003176 RVE40327.1 ABLF02017803 RSAL01000087 RVE48224.1 ABLF02014046 ABLF02014054 ABLF02014055 ABLF02015166 ABLF02066528 GEZM01101736 JAV52346.1 RSAL01005651 RVE39984.1 KQ971610 KYB24798.1 KQ972202 KYB24605.1 RSAL01000086 RVE48289.1 ABLF02028653 ABLF02048521 ABLF02061836 ABLF02064667 ABLF02007924 RSAL01000375 RVE42104.1 ABLF02023911 KQ971372 EFA10524.2 LBMM01000128 KMR04828.1 RSAL01000031 RVE51568.1 ABLF02008211 ABLF02008212 ABLF02008213 ABLF02011424 ABLF02042790 GGMS01010854 MBY80057.1 GEZM01085093 JAV60239.1 GEZM01085094 JAV60238.1 GEZM01085095 JAV60237.1 GEZM01090725 JAV57179.1 KQ973114 KXZ75664.1 RSAL01000165 RVE45277.1 GL769271 EFZ10565.1 GL438186 EFN69434.1 ABLF02005766 ABLF02032373 ABLF02032374 GEZM01085096 JAV60236.1 APGK01046392 APGK01046393 KB741054 ENN74456.1 GL764659 EFZ17305.1 GEZM01001982 JAV97423.1 GEZM01001983 JAV97421.1 NEVH01003762 PNF39995.1 PYGN01000122 PSN53542.1
KPJ07793.1 ODYU01011352 SOQ57041.1 ODYU01010582 SOQ55794.1 ABLF02008338 ABLF02016627 ABLF02063349 ABLF02008684 ABLF02008686 ABLF02008687 ABLF02011948 ABLF02019656 ABLF02019662 ABLF02041486 ABLF02009647 ABLF02009778 ABLF02009779 ABLF02025341 ABLF02042390 ABLF02017444 ODYU01002680 SOQ40482.1 NWSH01000959 PCG73380.1 ABLF02028679 ABLF02056730 ABLF02017692 ABLF02017693 RSAL01008431 RVE39798.1 ABLF02010326 ABLF02010327 GGMR01000046 MBY12665.1 ABLF02019592 ABLF02016553 ABLF02016555 ABLF02049531 ABLF02011758 ABLF02011759 ABLF02012658 LJIG01001987 KRT84953.1 ABLF02006763 ABLF02005638 ABLF02005639 ABLF02031840 ABLF02012220 ABLF02012222 ABLF02053218 GGMR01002126 MBY14745.1 GEZM01001796 JAV97521.1 GGMR01012308 MBY24927.1 GGMR01000178 MBY12797.1 GFXV01007611 MBW19416.1 JTDY01010331 KOB58244.1 GEZM01045715 JAV77678.1 NWSH01000363 PCG76969.1 PCG76967.1 PCG76968.1 LJIG01002596 KRT84344.1 ABLF02054892 RSAL01002057 RVE40596.1 ABLF02034357 ABLF02004413 ABLF02016452 ABLF02049812 ABLF02056148 ABLF02016989 JTDY01001960 KOB72446.1 RSAL01000885 RVE41073.1 GGMR01011939 MBY24558.1 ABLF02017696 RSAL01000699 RVE41218.1 ABLF02003651 ABLF02011697 RSAL01000257 RVE43416.1 ABLF02055473 ABLF02056006 GGMR01012897 MBY25516.1 RSAL01003176 RVE40327.1 ABLF02017803 RSAL01000087 RVE48224.1 ABLF02014046 ABLF02014054 ABLF02014055 ABLF02015166 ABLF02066528 GEZM01101736 JAV52346.1 RSAL01005651 RVE39984.1 KQ971610 KYB24798.1 KQ972202 KYB24605.1 RSAL01000086 RVE48289.1 ABLF02028653 ABLF02048521 ABLF02061836 ABLF02064667 ABLF02007924 RSAL01000375 RVE42104.1 ABLF02023911 KQ971372 EFA10524.2 LBMM01000128 KMR04828.1 RSAL01000031 RVE51568.1 ABLF02008211 ABLF02008212 ABLF02008213 ABLF02011424 ABLF02042790 GGMS01010854 MBY80057.1 GEZM01085093 JAV60239.1 GEZM01085094 JAV60238.1 GEZM01085095 JAV60237.1 GEZM01090725 JAV57179.1 KQ973114 KXZ75664.1 RSAL01000165 RVE45277.1 GL769271 EFZ10565.1 GL438186 EFN69434.1 ABLF02005766 ABLF02032373 ABLF02032374 GEZM01085096 JAV60236.1 APGK01046392 APGK01046393 KB741054 ENN74456.1 GL764659 EFZ17305.1 GEZM01001982 JAV97423.1 GEZM01001983 JAV97421.1 NEVH01003762 PNF39995.1 PYGN01000122 PSN53542.1
Proteomes
PRIDE
Pfam
Interpro
IPR021896
Transposase_37
+ More
IPR028073 PHTB1_N_dom
IPR036322 WD40_repeat_dom_sf
IPR026511 PTHB1
IPR028074 PHTB1_C_dom
IPR006612 THAP_Znf
IPR038441 THAP_Znf_sf
IPR025398 DUF4371
IPR008906 HATC_C_dom
IPR006816 ELMO_dom
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR021109 Peptidase_aspartic_dom_sf
IPR008737 Peptidase_asp_put
IPR005312 DUF1759
IPR005162 Retrotrans_gag_dom
IPR036361 SAP_dom_sf
IPR012337 RNaseH-like_sf
IPR003034 SAP_dom
IPR018289 MULE_transposase_dom
IPR013087 Znf_C2H2_type
IPR003593 AAA+_ATPase
IPR011527 ABC1_TM_dom
IPR027417 P-loop_NTPase
IPR036640 ABC1_TM_sf
IPR030244 Yor1
IPR003439 ABC_transporter-like
IPR017871 ABC_transporter_CS
IPR023378 YheA/YmcA-like_dom_sf
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR006580 Znf_TTF
IPR028073 PHTB1_N_dom
IPR036322 WD40_repeat_dom_sf
IPR026511 PTHB1
IPR028074 PHTB1_C_dom
IPR006612 THAP_Znf
IPR038441 THAP_Znf_sf
IPR025398 DUF4371
IPR008906 HATC_C_dom
IPR006816 ELMO_dom
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR021109 Peptidase_aspartic_dom_sf
IPR008737 Peptidase_asp_put
IPR005312 DUF1759
IPR005162 Retrotrans_gag_dom
IPR036361 SAP_dom_sf
IPR012337 RNaseH-like_sf
IPR003034 SAP_dom
IPR018289 MULE_transposase_dom
IPR013087 Znf_C2H2_type
IPR003593 AAA+_ATPase
IPR011527 ABC1_TM_dom
IPR027417 P-loop_NTPase
IPR036640 ABC1_TM_sf
IPR030244 Yor1
IPR003439 ABC_transporter-like
IPR017871 ABC_transporter_CS
IPR023378 YheA/YmcA-like_dom_sf
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR006580 Znf_TTF
SUPFAM
ProteinModelPortal
H9JIR1
F8SL48
A0A0N1IC55
A0A194QQL0
A0A2H1WVF6
A0A2H1WTP2
+ More
J9L1W2 X1WQI7 X1XTB8 J9M4P5 J9M961 J9LX62 J9L6Z3 J9KEP3 A0A2H1VJ77 A0A2A4JPP6 J9KEF2 J9M9N1 A0A3S2LLE6 J9LEP6 A0A2S2N697 J9M897 J9KSA4 X1XB84 J9LY75 A0A0T6BCB1 J9KBW7 X1XC48 J9LPQ0 J9KGM8 A0A2S2NC74 A0A1Y1NI49 A0A2S2P628 A0A2S2N6K3 A0A2H8TYS2 A0A0L7K5T4 A0A1Y1LY36 A0A2A4JYS8 A0A2A4JY71 A0A2A4JZ58 A0A0T6BAH8 X1XAX4 A0A3S2KX83 J9M6Y7 X1XMZ9 J9L3T7 A0A0L7LB47 A0A3S2LNK6 A0A2S2P5T9 J9L7T1 A0A437ASS2 J9KT02 X1XJT3 A0A3S2NSI9 J9KI00 A0A2S2P9C5 A0A3S2LM91 X1XGM9 A0A437BCW2 J9KFX0 A0A1Y1JZB1 A0A3S2NMS9 A0A139WA73 A0A139W9L5 A0A437BCX3 X1X0R9 J9L5B6 A0A3S2L9N1 J9KX16 D6X0C0 A0A0J7LAG3 A0A437BMU4 J9LQD8 A0A2S2QQP4 A0A1Y1KFJ5 A0A1Y1KI01 A0A1Y1KN14 A0A1Y1K6U6 A0A139W8P5 A0A3S2LEZ0 E9J9D7 E2AAV3 J9M7W9 A0A1Y1KLF8 N6U1G5 E9IPY0 A0A1Y1NHP2 A0A1Y1NHU8 A0A2J7RGL9 A0A2P8ZAL9
J9L1W2 X1WQI7 X1XTB8 J9M4P5 J9M961 J9LX62 J9L6Z3 J9KEP3 A0A2H1VJ77 A0A2A4JPP6 J9KEF2 J9M9N1 A0A3S2LLE6 J9LEP6 A0A2S2N697 J9M897 J9KSA4 X1XB84 J9LY75 A0A0T6BCB1 J9KBW7 X1XC48 J9LPQ0 J9KGM8 A0A2S2NC74 A0A1Y1NI49 A0A2S2P628 A0A2S2N6K3 A0A2H8TYS2 A0A0L7K5T4 A0A1Y1LY36 A0A2A4JYS8 A0A2A4JY71 A0A2A4JZ58 A0A0T6BAH8 X1XAX4 A0A3S2KX83 J9M6Y7 X1XMZ9 J9L3T7 A0A0L7LB47 A0A3S2LNK6 A0A2S2P5T9 J9L7T1 A0A437ASS2 J9KT02 X1XJT3 A0A3S2NSI9 J9KI00 A0A2S2P9C5 A0A3S2LM91 X1XGM9 A0A437BCW2 J9KFX0 A0A1Y1JZB1 A0A3S2NMS9 A0A139WA73 A0A139W9L5 A0A437BCX3 X1X0R9 J9L5B6 A0A3S2L9N1 J9KX16 D6X0C0 A0A0J7LAG3 A0A437BMU4 J9LQD8 A0A2S2QQP4 A0A1Y1KFJ5 A0A1Y1KI01 A0A1Y1KN14 A0A1Y1K6U6 A0A139W8P5 A0A3S2LEZ0 E9J9D7 E2AAV3 J9M7W9 A0A1Y1KLF8 N6U1G5 E9IPY0 A0A1Y1NHP2 A0A1Y1NHU8 A0A2J7RGL9 A0A2P8ZAL9
Ontologies
GO
PANTHER
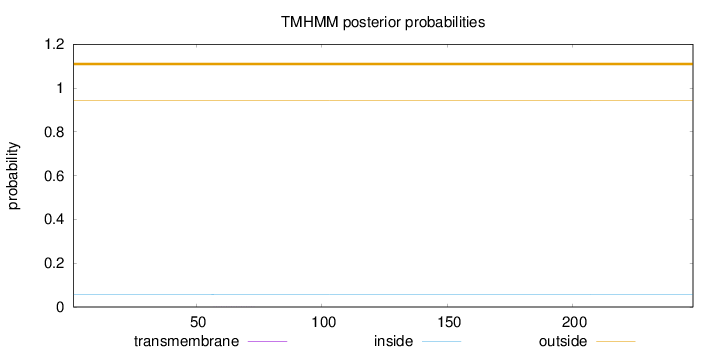
Topology
Length:
248
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00436
Exp number, first 60 AAs:
0.00164
Total prob of N-in:
0.05796
outside
1 - 248
Population Genetic Test Statistics
Pi
109.353919
Theta
169.695258
Tajima's D
-1.119427
CLR
244.457933
CSRT
0.115544222788861
Interpretation
Uncertain
