Pre Gene Modal
BGIBMGA009405
Annotation
PREDICTED:_myosin_light_chain_kinase?_smooth_muscle-like_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 3.142
Sequence
CDS
ATGCTCTCGGAAATCGGACGAGGCAAATTCGGAACAGTATACTTGTGCAGAGAAAAGAGCACGGGTCTGGAACTGGCCGCCAAGCTCGTCCTAGTGAACAGAAGGGACGAGAGACGCAACGTGGAACGGGAGGTGGACGTGATGAGGAGACTCCGGCACCCTCGACTGATCCAACTGTACGACGCTTACGACTGGGGGAAATATATGTGCGTCGTCCTTGAACTAATAACGGGAGGCGAGCTGTTCGAAAGAGTCATCGACGAAGATTTCGTTCTGACAGAGCGCGCCTGTACTGTGTTCATGCGACAGATCTGCGAGGGCATCGAGTTCGTTCACCGCCAGAATATACTCCACCTGGATCTGAAGCCCGAAAACATTCTCTGCCTAACGAAGACCGGTAACCGTATCAAGATTATAGACTTTGGTCTCGCCAGATTCTATGACCCGGAGAAGAAGCTGCAAGTGTTGTTCGGGACCCCGGAGTTCGTTGCCCCCGAGGTGGTCAACTTCGATCAGATCGGTTACGGCACTGACATGTGGTCCGTCGGAGTCATATGTTATGTCCTACTATCAGGACTGTCCCCGTTCATGGGAGAAACCGACATAGAGACAATGGCGAACGTGACTGTCGCAAAATATGACTTCGACGATGAAGCCTTCAATGAAATATCCGAAGACGCAAAGGACTTCATACGGAAACTGCTCGTGAAAGACAAAGAGTCACGGCCCAGCGCGGCGGAGTGTCTCCGGCACGCGTGGGTGGCGCCCCCGCCCGCGCGGAAACATGAACTCGACGTCACCAAGGACAACCTGCGCCTCTTCGTCGAGCGCTGGGGCGACCATCCCAACTCTCCTTACGTCTTCGACAACGAAGCGCACGAGATCACCTGCCTCGCTCTCTGCGAGCGGTCCTCCGTCGGAGGCTGCTCGCCCTCTCCTCGAAGCTCCCTCGCCACGTCCCTCACGAGCTCGCCCGACGGAGTCTTCGACGAGGACGACTTGGAGCCCCCTCCGCTCAGAGACACTACATTCCCCGCCAAGAGATACAATCCTGTGGAACGCCGCGCTTCCGACAGTTCGTGCTTTTTGCATAAAAAACACGACGTGTTCGTTCGAAAGAATTTGGCCGAGGAAATAAAAAAGCTATCGGATCACCTTTTCATGCTGTCCACGATGAACACGGATCTGACGAACAACAACAACAGCAGAGAGCTCGATAAAAACGTTACGGAAAATAAAGAAATTAAAGAAGAGAAACTGCCCAACGGGATTAAGAAAGAAGCAAAAGTGACAGTGACGAAAACGGTGACCAATGGAGACGCTAATAAAAAAGATAGAAGAATATTCACATCTAAATCGACTACGAAGATATCGACAGCGTTTCTCACAGACAAGGACTCCGAGAGGCCGATGACGAGCAAAATGCCGTGGGCCAAGTCGACCAGGTCGAAACGAGTCACCAACTTAAGCAGAGACGTCCCTGAACAACCGGTGGAGGTCCGGAACAGCTTCTTCCAGGAATTCGACAAAAGACGCGATAAAATCGACAAACTAGTAAACGGCGCGGAAAACGGCCGCCAAATTCCCTACAGAAGGGGAGATGAGAACAACGCTCATAGAACGAAAGACCTCTTGTTACACTTATTAGAAAAATGGGGGGAGAACGAGGATATCGAAGCGGAGAGCGACCGTCTCTCCGCCTCGTCTAGTCTGACGCATCCGTGGCTGATCCAATCGGCGCTCTGCACTGAACTCCACGTCACCAAGACGAAACTCAAGCGCTACGTCATCAAGAAACGTTGGGCCAAGGCCGTCGCCGCAGTCATAGCCCTCAAGAGAATGGGCGCCAAATTCGAGGACGTCCATGAGGATGGTAGTAACAAAAACGGAGAAAAATAA
Protein
MLSEIGRGKFGTVYLCREKSTGLELAAKLVLVNRRDERRNVEREVDVMRRLRHPRLIQLYDAYDWGKYMCVVLELITGGELFERVIDEDFVLTERACTVFMRQICEGIEFVHRQNILHLDLKPENILCLTKTGNRIKIIDFGLARFYDPEKKLQVLFGTPEFVAPEVVNFDQIGYGTDMWSVGVICYVLLSGLSPFMGETDIETMANVTVAKYDFDDEAFNEISEDAKDFIRKLLVKDKESRPSAAECLRHAWVAPPPARKHELDVTKDNLRLFVERWGDHPNSPYVFDNEAHEITCLALCERSSVGGCSPSPRSSLATSLTSSPDGVFDEDDLEPPPLRDTTFPAKRYNPVERRASDSSCFLHKKHDVFVRKNLAEEIKKLSDHLFMLSTMNTDLTNNNNSRELDKNVTENKEIKEEKLPNGIKKEAKVTVTKTVTNGDANKKDRRIFTSKSTTKISTAFLTDKDSERPMTSKMPWAKSTRSKRVTNLSRDVPEQPVEVRNSFFQEFDKRRDKIDKLVNGAENGRQIPYRRGDENNAHRTKDLLLHLLEKWGENEDIEAESDRLSASSSLTHPWLIQSALCTELHVTKTKLKRYVIKKRWAKAVAAVIALKRMGAKFEDVHEDGSNKNGEK
Summary
Uniprot
EMBL
RSAL01000059
RVE49707.1
JTDY01000124
KOB78724.1
ODYU01002686
SOQ40511.1
+ More
KQ458751 KPJ05301.1 AGBW02009237 OWR51285.1 NWSH01001996 PCG69487.1 KQ971315 EEZ97856.2 APGK01043663 KB741015 ENN75369.1 KB632237 ERL90442.1 KQ434886 KZC10153.1 AJWK01003907 AJWK01003908 AJVK01019335 AJVK01019336 GFDF01007099 JAV06985.1 LJIG01022740 KRT78962.1 KA646920 AFP61549.1 CCAG010002097
KQ458751 KPJ05301.1 AGBW02009237 OWR51285.1 NWSH01001996 PCG69487.1 KQ971315 EEZ97856.2 APGK01043663 KB741015 ENN75369.1 KB632237 ERL90442.1 KQ434886 KZC10153.1 AJWK01003907 AJWK01003908 AJVK01019335 AJVK01019336 GFDF01007099 JAV06985.1 LJIG01022740 KRT78962.1 KA646920 AFP61549.1 CCAG010002097
Proteomes
Pfam
PF00069 Pkinase
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
PDB
2X4F
E-value=6.25897e-74,
Score=708
Ontologies
GO
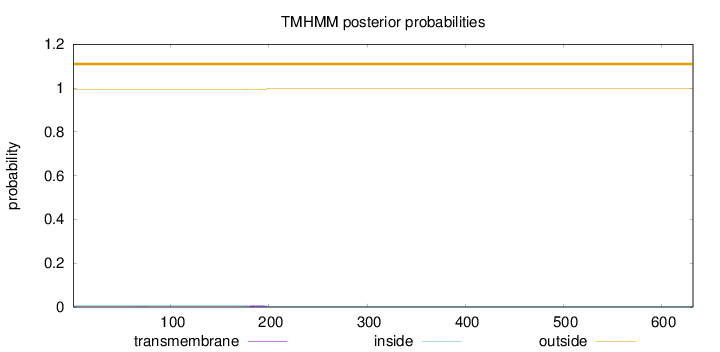
Topology
Length:
632
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.14885
Exp number, first 60 AAs:
0.00055
Total prob of N-in:
0.00718
outside
1 - 632
Population Genetic Test Statistics
Pi
245.182193
Theta
167.755861
Tajima's D
1.461226
CLR
0.41811
CSRT
0.772861356932153
Interpretation
Uncertain
