Pre Gene Modal
BGIBMGA009408
Annotation
PREDICTED:_DNA-directed_RNA_polymerase_III_subunit_RPC3_[Papilio_machaon]
Location in the cell
Cytoplasmic Reliability : 1.385 Mitochondrial Reliability : 2.006
Sequence
CDS
ATGTCACATCAGCTGGGCCGTGTTGTTTCTGTGATTCTCCAGAGATATTTTGGCGAGAATGTACAGAAAGTTGGCGACGATCTATTTACATATGGCAGTAAGCCCATTGCCTTATTAGTGAAGAGTACTGGTCTTTCCAGAATCAAGGTGGTAGAAAGTCTCCGAACGTTACTGAAGTATGATTTGGCAATGTTTGAACCAACTCCAAATGAAGTAATTGCAGATTACAGATTAATTCCGGAGAATGTGCTTTTGATGATTAGATATCCAAGGTATTTACTACAAATTAAAACGAAGTACGGTAGTGAAGCAGAAATATTGATTGAAGAGTTGTTACAAAAGGCATGTTCCACTGCTACCGTGCTATTAGTCTCCATTGCTGTTAAATATAAAGATGACAAGGAAAAGAACATGACTATAGTGAAACTAAAAGATATCTTTATAAGTCTGATAACCGCTGGTTACATACAGCAGGCATCTGTAGCTGAGTTGAAGGAAGGCAGCGAGATACCGACACTGGTGCCCGTCGCAACTATAACACCAGAGATTGATGTACGAGCTGTCATTAAAATTATGAATTCTGGAAATCTAGCTGAAAGTAGTGATAATGTGTACTGGAAAGTTAATTACGACAGGTTCCATCAGGATATCCGCGACGAAATTATGATCAAAGCAATCACAAGACGAATAGATGAAAACGCCGGTGAGCTTCTTCGTCATATGCTGCAACACATGTACCTGACGTCTCCGGCGTGGGCTGCAGAGTCTACACTAGTGCCGGCCGTGGAATTGAGGGAGGCTTGTCTCCGGGCCGCTGCTGACAAGCCCCTCTTGAATCAGTATTTCGATCAATATCTGAAGGTCTTAGAGGAGAGCGGGTGCGGCACGGTGCGGGCGGACGCGGGGGGGCGGGGGTACAGCGTGCGGGGGGCGGGCGCGGCGCGGGGGTGCCTGCTGGCGCTGCTGGAGGGGCTGGTCACGGAGAGACTGGGCTACAAGGCCGCCAGGATATTCAGAATAATTCTCGATAAGAAATATATAGAAGAGGATGACGTACAAAAAAACGCCATGCTACCAAATAAGGAATGCAAAGAGCTGTCGTATAAACTTTTAGAAGAGCATTTCATTAGTTTACAGCCAATGAGGAAAACCGCGTCGGCCGGCGGCATGAACAAGGCCGTCTATTTGTTCCATATTAATCTACACGACGTGGCACACACAGTGCAGGACATGTGCTACAAGGCTCTGGGCAACGTGTTGCGACGCGCGTCCCACGAGCGGGCGGCCCACGCGCGGCTGTTGGACAAGCAGCGCCGCGTGCGCAGCATAGTGCACGGCATGAGGCTGCGCGCCGAGCCGCAACAGAACATCGATGACGTTGAAGAAACGCTGACCCCGTCCGAACTAGCAGTACTGCAGGGCGTCGAGAAACGACTCAAACAGCTCAGCGCCGCTGAATTAGAAATCGATAAAAATCTATTTATACTGAAGTGGTACTTTATGTATACGCATTGA
Protein
MSHQLGRVVSVILQRYFGENVQKVGDDLFTYGSKPIALLVKSTGLSRIKVVESLRTLLKYDLAMFEPTPNEVIADYRLIPENVLLMIRYPRYLLQIKTKYGSEAEILIEELLQKACSTATVLLVSIAVKYKDDKEKNMTIVKLKDIFISLITAGYIQQASVAELKEGSEIPTLVPVATITPEIDVRAVIKIMNSGNLAESSDNVYWKVNYDRFHQDIRDEIMIKAITRRIDENAGELLRHMLQHMYLTSPAWAAESTLVPAVELREACLRAAADKPLLNQYFDQYLKVLEESGCGTVRADAGGRGYSVRGAGAARGCLLALLEGLVTERLGYKAARIFRIILDKKYIEEDDVQKNAMLPNKECKELSYKLLEEHFISLQPMRKTASAGGMNKAVYLFHINLHDVAHTVQDMCYKALGNVLRRASHERAAHARLLDKQRRVRSIVHGMRLRAEPQQNIDDVEETLTPSELAVLQGVEKRLKQLSAAELEIDKNLFILKWYFMYTH
Summary
Uniprot
H9JIQ7
A0A3S2NRN5
A0A0N1IMV8
A0A0N1IGL4
A0A212EPS1
D2A0B8
+ More
A0A2J7PPT7 A0A1B0GJ49 A0A1B0EZN9 A0A067RF68 A0A182G1I4 A0A182H0L2 Q171Y0 A0A182YKI1 A0A336MMU8 A0A1Y1M8V1 A0A1B6HFR0 A0A182VX58 A0A182NA87 A0A182MP67 A0A084W1P1 A0A336MJX9 B0WS61 U4UK77 N6TZ50 A0A154P4H2 A0A182J711 N6UQH7 A0A182R3N2 A0A182HT80 A0A182QQQ1 A0A182LFC2 A0A182TM07 A0A182V557 Q7QC11 N6U3E8 A0A182XI93 A0A182JX78 A0A182PA62 A0A087ZRQ3 A0A0L7R636 A0A1J1J894 A0A2M4ARR6 W5JHC4 A0A0L0CNV6 A0A182F8C9 A0A146KZX2 A0A0A9WE05 A0A0M9A4B3 A0A2A3EP69 B4PPB0 A0A1W4UN37 B4HGW7 B3P4A8 B4QS98 A0A232F5S4 B3M2S2 B4GZ76 A0A195F8J9 Q9VG60 K7J6S8 Q29B17 R4G8T2 A0A1I8MDW6 A0A3B0K5X2 A0A069DZ68 A0A1A9ZHC0 A0A1A9UCU3 A0A0V0G8I6 A0A1B0G3F3 B4KAA4 A0A0A1WLU6 A0A1I8P2D9 A0A0K8WFB9 A0A1A9Y8U7 A0A1B0BG50 F4WJY8 A0A034WCU0 A0A195BHZ1 A0A158N9D4 A0A1A9X417 B4M6A3 E2ANZ2 W8B7M2 B4JI82 A0A1B6GJ47 A0A026VTN7 A0A423U8Y7 B4N8R8 A0A1B6G3S8 A0A224XPV2 A0A195E6W0 A0A0M4ELL2 A0A1W4XJI8 A0A151XI39
A0A2J7PPT7 A0A1B0GJ49 A0A1B0EZN9 A0A067RF68 A0A182G1I4 A0A182H0L2 Q171Y0 A0A182YKI1 A0A336MMU8 A0A1Y1M8V1 A0A1B6HFR0 A0A182VX58 A0A182NA87 A0A182MP67 A0A084W1P1 A0A336MJX9 B0WS61 U4UK77 N6TZ50 A0A154P4H2 A0A182J711 N6UQH7 A0A182R3N2 A0A182HT80 A0A182QQQ1 A0A182LFC2 A0A182TM07 A0A182V557 Q7QC11 N6U3E8 A0A182XI93 A0A182JX78 A0A182PA62 A0A087ZRQ3 A0A0L7R636 A0A1J1J894 A0A2M4ARR6 W5JHC4 A0A0L0CNV6 A0A182F8C9 A0A146KZX2 A0A0A9WE05 A0A0M9A4B3 A0A2A3EP69 B4PPB0 A0A1W4UN37 B4HGW7 B3P4A8 B4QS98 A0A232F5S4 B3M2S2 B4GZ76 A0A195F8J9 Q9VG60 K7J6S8 Q29B17 R4G8T2 A0A1I8MDW6 A0A3B0K5X2 A0A069DZ68 A0A1A9ZHC0 A0A1A9UCU3 A0A0V0G8I6 A0A1B0G3F3 B4KAA4 A0A0A1WLU6 A0A1I8P2D9 A0A0K8WFB9 A0A1A9Y8U7 A0A1B0BG50 F4WJY8 A0A034WCU0 A0A195BHZ1 A0A158N9D4 A0A1A9X417 B4M6A3 E2ANZ2 W8B7M2 B4JI82 A0A1B6GJ47 A0A026VTN7 A0A423U8Y7 B4N8R8 A0A1B6G3S8 A0A224XPV2 A0A195E6W0 A0A0M4ELL2 A0A1W4XJI8 A0A151XI39
Pubmed
19121390
26354079
22118469
18362917
19820115
24845553
+ More
26483478 17510324 25244985 28004739 24438588 23537049 20966253 12364791 14747013 17210077 20920257 23761445 26108605 26823975 25401762 17994087 17550304 28648823 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20075255 15632085 25315136 26334808 25830018 21719571 25348373 21347285 20798317 24495485 24508170 30249741
26483478 17510324 25244985 28004739 24438588 23537049 20966253 12364791 14747013 17210077 20920257 23761445 26108605 26823975 25401762 17994087 17550304 28648823 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20075255 15632085 25315136 26334808 25830018 21719571 25348373 21347285 20798317 24495485 24508170 30249741
EMBL
BABH01032885
RSAL01000297
RVE42807.1
KQ459256
KPJ02071.1
KQ460426
+ More
KPJ14772.1 AGBW02013418 OWR43464.1 KQ971338 EFA02494.1 NEVH01022644 PNF18354.1 AJWK01019308 AJWK01019309 AJWK01019310 AJWK01019311 AJVK01007422 KK852672 KDR18768.1 JXUM01023447 KQ560661 KXJ81389.1 JXUM01101474 KQ564651 KXJ71958.1 CH477443 EAT40806.1 UFQS01001349 UFQT01001349 SSX10467.1 SSX30153.1 GEZM01037064 JAV82219.1 GECU01034139 JAS73567.1 AXCM01010392 ATLV01019399 KE525269 KFB44135.1 UFQS01002094 UFQT01001174 UFQT01002094 SSX13212.1 SSX29289.1 DS232065 EDS33684.1 KB632337 ERL92863.1 APGK01003070 KB735104 ENN83448.1 KQ434814 KZC06816.1 APGK01007669 KB737396 ENN83031.1 APCN01000421 AXCN02000218 AAAB01008859 EAA08103.3 APGK01041235 KB740992 ENN76090.1 KQ414648 KOC66231.1 CVRI01000070 CRL07129.1 GGFK01010143 MBW43464.1 ADMH02001252 ETN63461.1 JRES01000232 KNC33139.1 GDHC01016738 JAQ01891.1 GBHO01036922 GBRD01006522 JAG06682.1 JAG59299.1 KQ435737 KOX76947.1 KZ288202 PBC33528.1 CM000160 EDW97116.1 CH480815 EDW42438.1 CH954181 EDV49353.1 CM000364 EDX13194.1 NNAY01000856 OXU26186.1 CH902617 EDV42393.1 CH479198 EDW28094.1 KQ981727 KYN36701.1 AE014297 AY051718 AAF54828.2 AAK93142.1 AAZX01000215 CM000070 EAL27182.1 ACPB03006727 GAHY01000427 JAA77083.1 OUUW01000005 SPP81399.1 GBGD01001165 JAC87724.1 GECL01001787 JAP04337.1 CCAG010018501 CH933806 EDW14591.1 GBXI01015456 GBXI01014787 JAC98835.1 JAC99504.1 GDHF01002532 JAI49782.1 JXJN01013752 GL888187 EGI65510.1 GAKP01006453 JAC52499.1 KQ976476 KYM83850.1 ADTU01009481 CH940652 EDW59179.1 GL441439 EFN64824.1 GAMC01013511 GAMC01013510 GAMC01013507 JAB93048.1 CH916369 EDV92963.1 GECZ01007324 JAS62445.1 KK108118 QOIP01000005 EZA46881.1 RLU22415.1 QCYY01000426 ROT85163.1 CH964232 EDW81519.1 GECZ01012679 JAS57090.1 GFTR01006397 JAW10029.1 KQ979592 KYN20584.1 CP012526 ALC45251.1 ALC47182.1 KQ982093 KYQ60072.1
KPJ14772.1 AGBW02013418 OWR43464.1 KQ971338 EFA02494.1 NEVH01022644 PNF18354.1 AJWK01019308 AJWK01019309 AJWK01019310 AJWK01019311 AJVK01007422 KK852672 KDR18768.1 JXUM01023447 KQ560661 KXJ81389.1 JXUM01101474 KQ564651 KXJ71958.1 CH477443 EAT40806.1 UFQS01001349 UFQT01001349 SSX10467.1 SSX30153.1 GEZM01037064 JAV82219.1 GECU01034139 JAS73567.1 AXCM01010392 ATLV01019399 KE525269 KFB44135.1 UFQS01002094 UFQT01001174 UFQT01002094 SSX13212.1 SSX29289.1 DS232065 EDS33684.1 KB632337 ERL92863.1 APGK01003070 KB735104 ENN83448.1 KQ434814 KZC06816.1 APGK01007669 KB737396 ENN83031.1 APCN01000421 AXCN02000218 AAAB01008859 EAA08103.3 APGK01041235 KB740992 ENN76090.1 KQ414648 KOC66231.1 CVRI01000070 CRL07129.1 GGFK01010143 MBW43464.1 ADMH02001252 ETN63461.1 JRES01000232 KNC33139.1 GDHC01016738 JAQ01891.1 GBHO01036922 GBRD01006522 JAG06682.1 JAG59299.1 KQ435737 KOX76947.1 KZ288202 PBC33528.1 CM000160 EDW97116.1 CH480815 EDW42438.1 CH954181 EDV49353.1 CM000364 EDX13194.1 NNAY01000856 OXU26186.1 CH902617 EDV42393.1 CH479198 EDW28094.1 KQ981727 KYN36701.1 AE014297 AY051718 AAF54828.2 AAK93142.1 AAZX01000215 CM000070 EAL27182.1 ACPB03006727 GAHY01000427 JAA77083.1 OUUW01000005 SPP81399.1 GBGD01001165 JAC87724.1 GECL01001787 JAP04337.1 CCAG010018501 CH933806 EDW14591.1 GBXI01015456 GBXI01014787 JAC98835.1 JAC99504.1 GDHF01002532 JAI49782.1 JXJN01013752 GL888187 EGI65510.1 GAKP01006453 JAC52499.1 KQ976476 KYM83850.1 ADTU01009481 CH940652 EDW59179.1 GL441439 EFN64824.1 GAMC01013511 GAMC01013510 GAMC01013507 JAB93048.1 CH916369 EDV92963.1 GECZ01007324 JAS62445.1 KK108118 QOIP01000005 EZA46881.1 RLU22415.1 QCYY01000426 ROT85163.1 CH964232 EDW81519.1 GECZ01012679 JAS57090.1 GFTR01006397 JAW10029.1 KQ979592 KYN20584.1 CP012526 ALC45251.1 ALC47182.1 KQ982093 KYQ60072.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000053240
UP000007151
UP000007266
+ More
UP000235965 UP000092461 UP000092462 UP000027135 UP000069940 UP000249989 UP000008820 UP000076408 UP000075920 UP000075884 UP000075883 UP000030765 UP000002320 UP000030742 UP000019118 UP000076502 UP000075880 UP000075900 UP000075840 UP000075886 UP000075882 UP000075902 UP000075903 UP000007062 UP000076407 UP000075881 UP000075885 UP000005203 UP000053825 UP000183832 UP000000673 UP000037069 UP000069272 UP000053105 UP000242457 UP000002282 UP000192221 UP000001292 UP000008711 UP000000304 UP000215335 UP000007801 UP000008744 UP000078541 UP000000803 UP000002358 UP000001819 UP000015103 UP000095301 UP000268350 UP000092445 UP000078200 UP000092444 UP000009192 UP000095300 UP000092443 UP000092460 UP000007755 UP000078540 UP000005205 UP000091820 UP000008792 UP000000311 UP000001070 UP000053097 UP000279307 UP000283509 UP000007798 UP000078492 UP000092553 UP000192223 UP000075809
UP000235965 UP000092461 UP000092462 UP000027135 UP000069940 UP000249989 UP000008820 UP000076408 UP000075920 UP000075884 UP000075883 UP000030765 UP000002320 UP000030742 UP000019118 UP000076502 UP000075880 UP000075900 UP000075840 UP000075886 UP000075882 UP000075902 UP000075903 UP000007062 UP000076407 UP000075881 UP000075885 UP000005203 UP000053825 UP000183832 UP000000673 UP000037069 UP000069272 UP000053105 UP000242457 UP000002282 UP000192221 UP000001292 UP000008711 UP000000304 UP000215335 UP000007801 UP000008744 UP000078541 UP000000803 UP000002358 UP000001819 UP000015103 UP000095301 UP000268350 UP000092445 UP000078200 UP000092444 UP000009192 UP000095300 UP000092443 UP000092460 UP000007755 UP000078540 UP000005205 UP000091820 UP000008792 UP000000311 UP000001070 UP000053097 UP000279307 UP000283509 UP000007798 UP000078492 UP000092553 UP000192223 UP000075809
Interpro
SUPFAM
SSF47923
SSF47923
Gene 3D
ProteinModelPortal
H9JIQ7
A0A3S2NRN5
A0A0N1IMV8
A0A0N1IGL4
A0A212EPS1
D2A0B8
+ More
A0A2J7PPT7 A0A1B0GJ49 A0A1B0EZN9 A0A067RF68 A0A182G1I4 A0A182H0L2 Q171Y0 A0A182YKI1 A0A336MMU8 A0A1Y1M8V1 A0A1B6HFR0 A0A182VX58 A0A182NA87 A0A182MP67 A0A084W1P1 A0A336MJX9 B0WS61 U4UK77 N6TZ50 A0A154P4H2 A0A182J711 N6UQH7 A0A182R3N2 A0A182HT80 A0A182QQQ1 A0A182LFC2 A0A182TM07 A0A182V557 Q7QC11 N6U3E8 A0A182XI93 A0A182JX78 A0A182PA62 A0A087ZRQ3 A0A0L7R636 A0A1J1J894 A0A2M4ARR6 W5JHC4 A0A0L0CNV6 A0A182F8C9 A0A146KZX2 A0A0A9WE05 A0A0M9A4B3 A0A2A3EP69 B4PPB0 A0A1W4UN37 B4HGW7 B3P4A8 B4QS98 A0A232F5S4 B3M2S2 B4GZ76 A0A195F8J9 Q9VG60 K7J6S8 Q29B17 R4G8T2 A0A1I8MDW6 A0A3B0K5X2 A0A069DZ68 A0A1A9ZHC0 A0A1A9UCU3 A0A0V0G8I6 A0A1B0G3F3 B4KAA4 A0A0A1WLU6 A0A1I8P2D9 A0A0K8WFB9 A0A1A9Y8U7 A0A1B0BG50 F4WJY8 A0A034WCU0 A0A195BHZ1 A0A158N9D4 A0A1A9X417 B4M6A3 E2ANZ2 W8B7M2 B4JI82 A0A1B6GJ47 A0A026VTN7 A0A423U8Y7 B4N8R8 A0A1B6G3S8 A0A224XPV2 A0A195E6W0 A0A0M4ELL2 A0A1W4XJI8 A0A151XI39
A0A2J7PPT7 A0A1B0GJ49 A0A1B0EZN9 A0A067RF68 A0A182G1I4 A0A182H0L2 Q171Y0 A0A182YKI1 A0A336MMU8 A0A1Y1M8V1 A0A1B6HFR0 A0A182VX58 A0A182NA87 A0A182MP67 A0A084W1P1 A0A336MJX9 B0WS61 U4UK77 N6TZ50 A0A154P4H2 A0A182J711 N6UQH7 A0A182R3N2 A0A182HT80 A0A182QQQ1 A0A182LFC2 A0A182TM07 A0A182V557 Q7QC11 N6U3E8 A0A182XI93 A0A182JX78 A0A182PA62 A0A087ZRQ3 A0A0L7R636 A0A1J1J894 A0A2M4ARR6 W5JHC4 A0A0L0CNV6 A0A182F8C9 A0A146KZX2 A0A0A9WE05 A0A0M9A4B3 A0A2A3EP69 B4PPB0 A0A1W4UN37 B4HGW7 B3P4A8 B4QS98 A0A232F5S4 B3M2S2 B4GZ76 A0A195F8J9 Q9VG60 K7J6S8 Q29B17 R4G8T2 A0A1I8MDW6 A0A3B0K5X2 A0A069DZ68 A0A1A9ZHC0 A0A1A9UCU3 A0A0V0G8I6 A0A1B0G3F3 B4KAA4 A0A0A1WLU6 A0A1I8P2D9 A0A0K8WFB9 A0A1A9Y8U7 A0A1B0BG50 F4WJY8 A0A034WCU0 A0A195BHZ1 A0A158N9D4 A0A1A9X417 B4M6A3 E2ANZ2 W8B7M2 B4JI82 A0A1B6GJ47 A0A026VTN7 A0A423U8Y7 B4N8R8 A0A1B6G3S8 A0A224XPV2 A0A195E6W0 A0A0M4ELL2 A0A1W4XJI8 A0A151XI39
PDB
5AFQ
E-value=5.15683e-56,
Score=552
Ontologies
GO
PANTHER
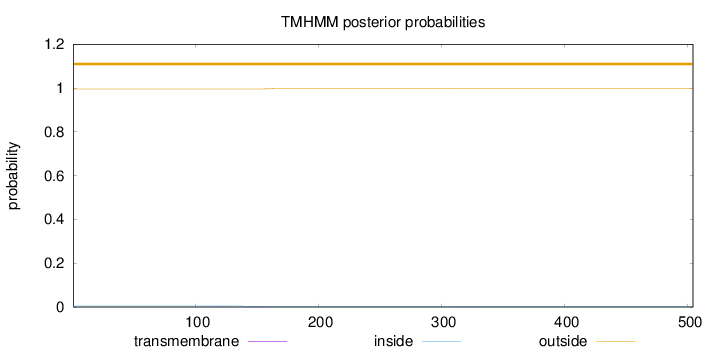
Topology
Length:
504
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0487400000000002
Exp number, first 60 AAs:
0.00042
Total prob of N-in:
0.00422
outside
1 - 504
Population Genetic Test Statistics
Pi
300.01948
Theta
190.192861
Tajima's D
1.934754
CLR
13.040873
CSRT
0.872006399680016
Interpretation
Uncertain
