Pre Gene Modal
BGIBMGA009409
Annotation
PREDICTED:_cleavage_and_polyadenylation_specificity_factor_subunit_CG7185_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.597
Sequence
CDS
ATGGCGGACGGCGATGTCGATATAGATTTGTACGCCGATGATATAGAATCCGATTTTAATCGCCAGGATGAATTTGCTGGCGAAAATGTGGATTTATATGACGACGTGATCTCAGCACCAGCTTCGAAGCCCGAGGACGGTGACGGTCCTCCCATCATTGCTCCAAACCAGCATCCACCTGAGGAAACTAACGGCAACGCCTCATACCACGGCCCGGTTCACGGACATCATGGCCGCCGTTTTCAACTTTACGTCGGCAACCTAACATGGTGGGCAACGGACCAAGATATTGCGAACGCGATCTCGGATGTCGGTGTCTCCGATTTTCAAGATGTGAAGTTCTTTGAGAACAGAGCCAATGGTCAGTCCAAAGGATTCTGCGTTATATCTCTCGGTTCAGATCAATCAGTCAGGATGGTGATGGAACGTCTTTCTAAGAAGGAGATACACGGGCAACATCCAGTAGTTACATTGCCTACTAAACAGGCGTTGAATCAGTTTGAAAGTCAGTCAAAGACCAGGTCGACGCCTCCTGGTCCCAATAATCCTGGAATGAGAGGACCCCATCCAGGCGGGCCGCCTGGTCCTCATCCCGGAGAATTTTTTGGTGGGGGACCGAACGGTCCAGGTGGGGGCGGACCTCGTATGATGCTCCCAGGTCCGGGTCCGCACCAGCTTCGTGGGCCGCCGCCGCACCACATGGGCCCACACCAGGGCGGGCCGCATCCCATGCAACATCAGCCGCAGCCGCACCAGGCCCCGCCACACCGGCCCCCTATGCAGTTTGCGGGCCCTCCGCAGATGCAACGCGCGCCGGGCCCGCGTCCGCCGCCTCCCGACTGGGCTGCGCGAGCGCCGCATCCTATGTCTCAGCCCGCGCCTCATTATGGACCCCCCCATCATGGAATGCCGCATCAGATGCCACCACCTCATCCTCAACCCGGCGCACAGGGCATGCCGCCTCCGCACCAGTTGCCGCCGCACCAGCCGCCGCGGGGACCGGCGCCCATGCCGCAGCATCCTGTAATACCTGGAGCTCCAGCACCACACGTGAATCCTGCGTTCTTCAACCAGCAGCCACCGTCGCAGCCGCCGCCAATGCAGGCCCCGCAACCTGGACCACCCGGTCCAGGTGCTCCGTACGGTCATCCAGCCCATGGAGCACCACCTCCAGCACAAGGGCCTCCACCCGGGTCTAGACAACCGTACGGTCGGCCACCACAGAGCTATCCGCCTCAAGCGGATCCGGCTCGTCCCCCGCACCACCCGGCACCCATGGTCCCACAGCACGCGGCCTCCCTGGCGCCCCCCATGCCGGTCATCAGCGAGGCCGAGTTCGAAGAGGTGATGAGCAGGAACCGCACCGTCTCCTCCAGTGCCATTGCTCGGGCTGTCAGTGACGCTGCCGCTGGCGAGTACGCCTCCGCCATCGAGACGCTCGTTACCGCCATATCACTCATTAAACAGAGCAAGGTGGCGCAAGACGACCGCTGCAAGATTCTGATCTCGTCGCTGCAAGACACGCTGCACGGCGTCGAGACGAAGTCTTACAGCAGCGGCGAGCGGTCCCGCCGCTCCCGGTCCCGCGAGCGCGAGCCGCGACACCACCGCGCCCCGCGCCGCCGCGACCGCTCTTCCAGCCGCTACCGGGACCGCAGCCGCGACCGAGAGGATAGGGACAGGTACAGGGAACGGTCGAGAGATCGCGACCGCGAACGTGAACGTGACGCGTATTACAGGGATTACCGTGAGAGGGAGCGCGAGAGGTCTCGCTCGAGGGATCGCGATCGTGCTGATCATTACAACAGGGGACACAGTCGTGAGGAGAGGGCACGTAAATCTCCAGCTGAAGCGACAGCAGAGGGAACCACAGCCGAGGCCGGGTCGAAGAGCCGCAGCACTGGAGCGCCGTACTACGACGAGCGGTACAGGGAGAGACGCGACAGAGACCCGCCGCCCGCGGATCGAGATCGCGACCGAGACCATCGCAGGGACGCTAGACACTAG
Protein
MADGDVDIDLYADDIESDFNRQDEFAGENVDLYDDVISAPASKPEDGDGPPIIAPNQHPPEETNGNASYHGPVHGHHGRRFQLYVGNLTWWATDQDIANAISDVGVSDFQDVKFFENRANGQSKGFCVISLGSDQSVRMVMERLSKKEIHGQHPVVTLPTKQALNQFESQSKTRSTPPGPNNPGMRGPHPGGPPGPHPGEFFGGGPNGPGGGGPRMMLPGPGPHQLRGPPPHHMGPHQGGPHPMQHQPQPHQAPPHRPPMQFAGPPQMQRAPGPRPPPPDWAARAPHPMSQPAPHYGPPHHGMPHQMPPPHPQPGAQGMPPPHQLPPHQPPRGPAPMPQHPVIPGAPAPHVNPAFFNQQPPSQPPPMQAPQPGPPGPGAPYGHPAHGAPPPAQGPPPGSRQPYGRPPQSYPPQADPARPPHHPAPMVPQHAASLAPPMPVISEAEFEEVMSRNRTVSSSAIARAVSDAAAGEYASAIETLVTAISLIKQSKVAQDDRCKILISSLQDTLHGVETKSYSSGERSRRSRSREREPRHHRAPRRRDRSSSRYRDRSRDREDRDRYRERSRDRDRERERDAYYRDYRERERERSRSRDRDRADHYNRGHSREERARKSPAEATAEGTTAEAGSKSRSTGAPYYDERYRERRDRDPPPADRDRDRDHRRDARH
Summary
Uniprot
ProteinModelPortal
PDB
3Q2T
E-value=3.14237e-32,
Score=348
Ontologies
GO
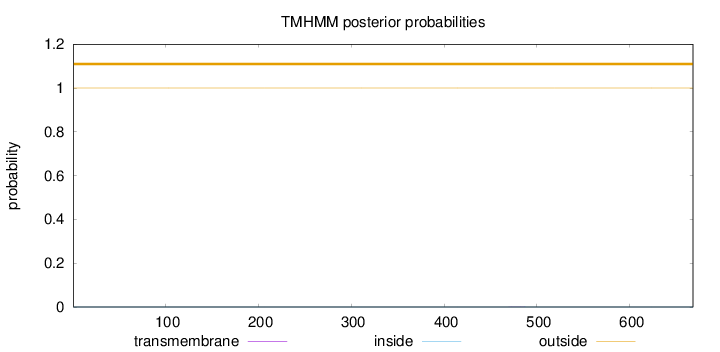
Topology
Length:
668
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00265
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00025
outside
1 - 668
Population Genetic Test Statistics
Pi
252.003729
Theta
173.240747
Tajima's D
1.349975
CLR
0.192123
CSRT
0.747162641867907
Interpretation
Uncertain
