Gene
KWMTBOMO08429
Annotation
PREDICTED:_intracellular_protein_transport_protein_USO1-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.29
Sequence
CDS
ATGGAGGTGCAAGAAAAATACGACTGGTTAGAAGGAGCAATCAAAGCAGCTATTCAACAAGGGACTAATAACAAAGACCTATCTAACAAGTGGATGACTACCAAAACAATCAACCTTCTCAAACAAAGAGCTAACTTAATTAAAGAAAAAAATAGTAACGATAATCGAAAGCAAATAGCAAAACTCAGCAAAGAAATTAAAGAAAGTATTAGAAAAGATAGAACTCAAAAAAGAATGGAAACAATAGAGAGACACATTTTACAAACAGGAGGTATAAAAAAGGCACATAAGGAATTATCAAACAAAAAGGACTGGATCGTACAGATGAAAGATAGTAATAGCTTGAAGGAAAATCGAAGGCAAGGAATATTAGGAATTGCTAGATCTTACTACAAGAAATTATATGAAAACAACATAGAGGAAAAAGAAATAGAGTTGTTGGAATTCTCATCCGTGCCCAGCATCATACAGGATGAAATAGAATTCGCTATAGAAACTCAAAGAGACAATAAAGCTCCAGGACCCGACAGAATAAGTAACGAAGTACTAAAACAAGCGAAACAAACAATAACACCTATTCTCAAAGAAATCTTCAACGATATCATCAACACAGAAACGATTCCCCAGCAATGGACTAAATCCAATATAATCCTTCTGTACAAAAAAGTATAA
Protein
MEVQEKYDWLEGAIKAAIQQGTNNKDLSNKWMTTKTINLLKQRANLIKEKNSNDNRKQIAKLSKEIKESIRKDRTQKRMETIERHILQTGGIKKAHKELSNKKDWIVQMKDSNSLKENRRQGILGIARSYYKKLYENNIEEKEIELLEFSSVPSIIQDEIEFAIETQRDNKAPGPDRISNEVLKQAKQTITPILKEIFNDIINTETIPQQWTKSNIILLYKKV
Summary
Uniprot
D7F179
A0A2W1BCS2
D7F176
A0A0L7L586
A0A2W1BNL4
A0A0L7K3E9
+ More
A0A0L7L0N7 A0A0L7K3E2 D7F177 A0A023EXV2 A0A2H1WF27 A0A2W1BZV1 A0A2W1BGR8 A0A212FGG8 A0A0N5BPB7 A0A0N4W1J5 A0A0K0FAW8 T1HR22 A0A2W1B5F8 A0A2H1VBB6 A0A0L7K349 A0A0N5C0B1 A0A0L7LUV7 A0A0L7K2L1 A0A3P9HFJ3 A0A069DZM5 A0A0L7KTL6 A0A3P9LQX3 A0A016WIQ2 A0A0K0D495 A0A3D1L484 F1KYC2 A0A016VCK8 A0A016V6A2 A0A016UN79 A0A016T4J8 A0A016T590 A0A016TWQ1 A0A016X2C0 A0A016SPR9 A0A016VEC1 A0A016S403 A0A016U784 A0A016VCD9 W6NY38 A0A016VCU3 A0A016WCB8 A0A016ULU9 A0A016WPY4 A0A016VBP7 A0A016S2X6 G0MPL1 D7F157 D7F163 A0A016U9D0 A0A016W940 A0A016THC6 A0A016USW0 A0A016UKG5 A0A016WA86 A0A016SSP6 A0A016UC68 A0A016SBW4 A0A016SCW1 A0A016SGC3 D7F164 A0A016VX03 A0A016TL37 A0A016W5P9 W6NFR9 A0A016W4R1 A0A2H1VS24
A0A0L7L0N7 A0A0L7K3E2 D7F177 A0A023EXV2 A0A2H1WF27 A0A2W1BZV1 A0A2W1BGR8 A0A212FGG8 A0A0N5BPB7 A0A0N4W1J5 A0A0K0FAW8 T1HR22 A0A2W1B5F8 A0A2H1VBB6 A0A0L7K349 A0A0N5C0B1 A0A0L7LUV7 A0A0L7K2L1 A0A3P9HFJ3 A0A069DZM5 A0A0L7KTL6 A0A3P9LQX3 A0A016WIQ2 A0A0K0D495 A0A3D1L484 F1KYC2 A0A016VCK8 A0A016V6A2 A0A016UN79 A0A016T4J8 A0A016T590 A0A016TWQ1 A0A016X2C0 A0A016SPR9 A0A016VEC1 A0A016S403 A0A016U784 A0A016VCD9 W6NY38 A0A016VCU3 A0A016WCB8 A0A016ULU9 A0A016WPY4 A0A016VBP7 A0A016S2X6 G0MPL1 D7F157 D7F163 A0A016U9D0 A0A016W940 A0A016THC6 A0A016USW0 A0A016UKG5 A0A016WA86 A0A016SSP6 A0A016UC68 A0A016SBW4 A0A016SCW1 A0A016SGC3 D7F164 A0A016VX03 A0A016TL37 A0A016W5P9 W6NFR9 A0A016W4R1 A0A2H1VS24
Pubmed
EMBL
FJ265564
ADI61832.1
KZ150485
PZC70710.1
FJ265561
ADI61829.1
+ More
JTDY01002915 KOB70496.1 KZ150049 PZC74456.1 JTDY01012202 KOB52316.1 JTDY01003910 KOB68839.1 JTDY01014598 KOB52012.1 FJ265562 ADI61830.1 GBBI01004876 JAC13836.1 ODYU01008249 SOQ51673.1 KZ149907 PZC78256.1 KZ150191 PZC72397.1 AGBW02008673 OWR52827.1 UZAF01016124 VDO21057.1 ACPB03015413 KZ150317 PZC71439.1 ODYU01001637 SOQ38140.1 JTDY01016396 KOB51897.1 JTDY01000076 KOB79001.1 JTDY01014306 KOB52041.1 GBGD01000380 JAC88509.1 JTDY01005938 KOB66455.1 JARK01000254 EYC39481.1 DPCO01000033 HCE59365.1 JI167974 ADY42876.1 JARK01001348 EYC25145.1 JARK01001353 EYC22263.1 JARK01001370 EYC16376.1 JARK01001471 EYB97918.1 JARK01001473 EYB97806.1 JARK01001407 EYC07235.1 JARK01000011 EYC46025.1 JARK01001531 EYB92324.1 EYC25048.1 JARK01001639 EYB85201.1 JARK01001389 EYC11015.1 EYC25050.1 CAVP010061281 CDL96712.1 EYC25046.1 JARK01000388 EYC37469.1 EYC16150.1 JARK01000173 EYC41342.1 EYC25049.1 JARK01001647 EYB84687.1 GL379805 EGT39774.1 FJ265542 ADI61810.1 FJ265548 ADI61816.1 JARK01001387 EYC11218.1 JARK01000575 EYC35812.1 JARK01001438 EYC02111.1 JARK01001365 EYC18011.1 JARK01001373 EYC15347.1 JARK01000460 EYC36759.1 JARK01001517 EYB93530.1 JARK01001383 EYC12427.1 JARK01001588 EYB88110.1 EYB88109.1 JARK01001568 EYB89432.1 FJ265549 ADI61817.1 JARK01001340 EYC31303.1 JARK01001430 EYC03362.1 JARK01000732 EYC35149.1 CAVP010059184 CAVP010059185 CDL95610.1 CDL95611.1 JARK01001337 EYC34625.1 ODYU01004088 SOQ43598.1
JTDY01002915 KOB70496.1 KZ150049 PZC74456.1 JTDY01012202 KOB52316.1 JTDY01003910 KOB68839.1 JTDY01014598 KOB52012.1 FJ265562 ADI61830.1 GBBI01004876 JAC13836.1 ODYU01008249 SOQ51673.1 KZ149907 PZC78256.1 KZ150191 PZC72397.1 AGBW02008673 OWR52827.1 UZAF01016124 VDO21057.1 ACPB03015413 KZ150317 PZC71439.1 ODYU01001637 SOQ38140.1 JTDY01016396 KOB51897.1 JTDY01000076 KOB79001.1 JTDY01014306 KOB52041.1 GBGD01000380 JAC88509.1 JTDY01005938 KOB66455.1 JARK01000254 EYC39481.1 DPCO01000033 HCE59365.1 JI167974 ADY42876.1 JARK01001348 EYC25145.1 JARK01001353 EYC22263.1 JARK01001370 EYC16376.1 JARK01001471 EYB97918.1 JARK01001473 EYB97806.1 JARK01001407 EYC07235.1 JARK01000011 EYC46025.1 JARK01001531 EYB92324.1 EYC25048.1 JARK01001639 EYB85201.1 JARK01001389 EYC11015.1 EYC25050.1 CAVP010061281 CDL96712.1 EYC25046.1 JARK01000388 EYC37469.1 EYC16150.1 JARK01000173 EYC41342.1 EYC25049.1 JARK01001647 EYB84687.1 GL379805 EGT39774.1 FJ265542 ADI61810.1 FJ265548 ADI61816.1 JARK01001387 EYC11218.1 JARK01000575 EYC35812.1 JARK01001438 EYC02111.1 JARK01001365 EYC18011.1 JARK01001373 EYC15347.1 JARK01000460 EYC36759.1 JARK01001517 EYB93530.1 JARK01001383 EYC12427.1 JARK01001588 EYB88110.1 EYB88109.1 JARK01001568 EYB89432.1 FJ265549 ADI61817.1 JARK01001340 EYC31303.1 JARK01001430 EYC03362.1 JARK01000732 EYC35149.1 CAVP010059184 CAVP010059185 CDL95610.1 CDL95611.1 JARK01001337 EYC34625.1 ODYU01004088 SOQ43598.1
Proteomes
Pfam
Interpro
Gene 3D
ProteinModelPortal
D7F179
A0A2W1BCS2
D7F176
A0A0L7L586
A0A2W1BNL4
A0A0L7K3E9
+ More
A0A0L7L0N7 A0A0L7K3E2 D7F177 A0A023EXV2 A0A2H1WF27 A0A2W1BZV1 A0A2W1BGR8 A0A212FGG8 A0A0N5BPB7 A0A0N4W1J5 A0A0K0FAW8 T1HR22 A0A2W1B5F8 A0A2H1VBB6 A0A0L7K349 A0A0N5C0B1 A0A0L7LUV7 A0A0L7K2L1 A0A3P9HFJ3 A0A069DZM5 A0A0L7KTL6 A0A3P9LQX3 A0A016WIQ2 A0A0K0D495 A0A3D1L484 F1KYC2 A0A016VCK8 A0A016V6A2 A0A016UN79 A0A016T4J8 A0A016T590 A0A016TWQ1 A0A016X2C0 A0A016SPR9 A0A016VEC1 A0A016S403 A0A016U784 A0A016VCD9 W6NY38 A0A016VCU3 A0A016WCB8 A0A016ULU9 A0A016WPY4 A0A016VBP7 A0A016S2X6 G0MPL1 D7F157 D7F163 A0A016U9D0 A0A016W940 A0A016THC6 A0A016USW0 A0A016UKG5 A0A016WA86 A0A016SSP6 A0A016UC68 A0A016SBW4 A0A016SCW1 A0A016SGC3 D7F164 A0A016VX03 A0A016TL37 A0A016W5P9 W6NFR9 A0A016W4R1 A0A2H1VS24
A0A0L7L0N7 A0A0L7K3E2 D7F177 A0A023EXV2 A0A2H1WF27 A0A2W1BZV1 A0A2W1BGR8 A0A212FGG8 A0A0N5BPB7 A0A0N4W1J5 A0A0K0FAW8 T1HR22 A0A2W1B5F8 A0A2H1VBB6 A0A0L7K349 A0A0N5C0B1 A0A0L7LUV7 A0A0L7K2L1 A0A3P9HFJ3 A0A069DZM5 A0A0L7KTL6 A0A3P9LQX3 A0A016WIQ2 A0A0K0D495 A0A3D1L484 F1KYC2 A0A016VCK8 A0A016V6A2 A0A016UN79 A0A016T4J8 A0A016T590 A0A016TWQ1 A0A016X2C0 A0A016SPR9 A0A016VEC1 A0A016S403 A0A016U784 A0A016VCD9 W6NY38 A0A016VCU3 A0A016WCB8 A0A016ULU9 A0A016WPY4 A0A016VBP7 A0A016S2X6 G0MPL1 D7F157 D7F163 A0A016U9D0 A0A016W940 A0A016THC6 A0A016USW0 A0A016UKG5 A0A016WA86 A0A016SSP6 A0A016UC68 A0A016SBW4 A0A016SCW1 A0A016SGC3 D7F164 A0A016VX03 A0A016TL37 A0A016W5P9 W6NFR9 A0A016W4R1 A0A2H1VS24
Ontologies
PANTHER
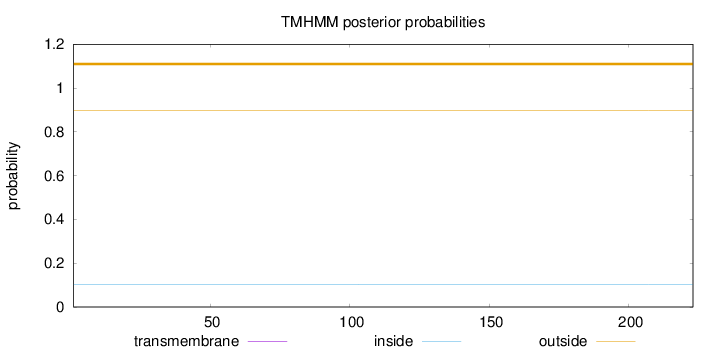
Topology
Length:
223
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.10359
outside
1 - 223
Population Genetic Test Statistics
Pi
3.323754
Theta
17.47468
Tajima's D
0
CLR
72.83979
CSRT
0.370581470926454
Interpretation
Uncertain
