Gene
KWMTBOMO08425
Pre Gene Modal
BGIBMGA009303
Annotation
PREDICTED:_synaptotagmin-10-like_isoform_X1_[Papilio_polytes]
Full name
Synaptotagmin-6
Alternative Name
Synaptotagmin VI
Location in the cell
Nuclear Reliability : 2.733
Sequence
CDS
ATGCCAGGATTGTGGCAACCGGAGAGTTTGGAAGGTCATACATCTGTGGTCGGGAGTTCCGTGGACTACGCAGCCGCGGTGCTACTGGCTGCGATGGCGGCTGTGTTATTAGCTGCTGTCGGTTACCAGTGCGCCGCTACTTGGAGATGGATCATGGGCAGGACTCGACGTGCCTCCCAGGAGAGCAACTGCGACTCCCTATCCCGCCAGGCAGCCATCCGCATTAGCCACTCTCTACCCGATCTGCAGACCGAGCCCCTGAAGCAGGAATATGTACAAGAGCACAAGGACCCAGCAAAGAAAGTCCTCCGTCAGACAACCCTGCCGATAGTTCCAACTCGCCACCAGACCTTTCAACGTCAGCTATCACACCGTCTGGAACTGCCCTCGATTCAGTTTAGCATCTGCAGTCTGGAACGTGCTGACTCGTCCATTGGGCTTATCAAGCCGGAGCTTTATAAGAACGAGTTGGTTCGTCAGTCGTCGACTGATAGTGCCGAATTGGAATTCTGCGGGAAGCTACACTTCGCTTTAAGATACGACCAGGAGGTTGAGGCCCTGATTGTAAAGATCTTCGAAGCTCGTGACCTACCGATTAAAGACATGAGCGGTAGCAGTGATCCATACATCAAGGTATTCCTCCTTCCCGACCGCAAGAAGAAGTTTCAGACGAAGGTCCACCGCAAGAACTTGAATCCGGTTTTCAATGAGACCTTTCTCTTCAGTGTACCGTACGAGGAGCTGCGCCAACGTTATCTGCAGTTTTCCGTGTACGATTTCGACCGGTTCAGCCGTCATGATCTGATCGGGCATGTTGTGCTGAAGGGACTACTGGAATCCGCTGACTTGGATCAGGAGATCGAATACACAATGAACATATTGGCTGCACCTCAGGAGAAGAAGGATCTTGGTGAACTAATGCTGTCCTTGTGCTACTTGCCCACTGCTGGTCGTCTAACTGTCACCGTCATCAAAGGCAGGAATCTGAAGGCGATGGATATTAATGGATCTTCCGATCCATACGTGAAGATCTGCCTGATTTGCCAGAACAAGCGCATTAAAAAGAAGAAGACCACAGTTAAGAAGAATACCCTGTGCCCGGTGTACAATGAAGCTCTCGTCTTTGATCTACCGTCTGAGAACGTCTTCGATGTGACTCTGCTCGTAAAAGTTATCGACTATGATAGACTCAGCCCCAACGAATTGATCGGATGCACGGCCATCGGGTCGTCTTTGATTGGAATCGGCCGCGACCATTGGCTGGAGATGCTCGACAACCCCCGCAAGCCGGTCGCCCAGTGGTATCCTCTAAACAAAAGTCTACCAACTAACGTCTCCCAACCAAACAAAAATGATCAAAATGCCGGTCTGAGCTGTTTAAATGGAAGATGA
Protein
MPGLWQPESLEGHTSVVGSSVDYAAAVLLAAMAAVLLAAVGYQCAATWRWIMGRTRRASQESNCDSLSRQAAIRISHSLPDLQTEPLKQEYVQEHKDPAKKVLRQTTLPIVPTRHQTFQRQLSHRLELPSIQFSICSLERADSSIGLIKPELYKNELVRQSSTDSAELEFCGKLHFALRYDQEVEALIVKIFEARDLPIKDMSGSSDPYIKVFLLPDRKKKFQTKVHRKNLNPVFNETFLFSVPYEELRQRYLQFSVYDFDRFSRHDLIGHVVLKGLLESADLDQEIEYTMNILAAPQEKKDLGELMLSLCYLPTAGRLTVTVIKGRNLKAMDINGSSDPYVKICLICQNKRIKKKKTTVKKNTLCPVYNEALVFDLPSENVFDVTLLVKVIDYDRLSPNELIGCTAIGSSLIGIGRDHWLEMLDNPRKPVAQWYPLNKSLPTNVSQPNKNDQNAGLSCLNGR
Summary
Description
May be involved in Ca(2+)-dependent exocytosis of secretory vesicles through Ca(2+) and phospholipid binding to the C2 domain or may serve as Ca(2+) sensors in the process of vesicular trafficking and exocytosis. May mediate Ca(2+)-regulation of exocytosis in acrosomal reaction in sperm (By similarity).
Cofactor
Ca(2+)
Subunit
Isoform 1: Homodimer; disulfide-linked via the cysteine motif. Isoform 1: Can also form heterodimers with SYT3, SYT7, SYT9 and SYT10. Isoform 1: Interacts with STX1A, STX1B and STX2; the interaction is Ca(2+)-dependent. Isoform 2: Is not able to form homodimer and heterodimers.
Similarity
Belongs to the synaptotagmin family.
Keywords
Alternative splicing
Calcium
Cell junction
Cell membrane
Complete proteome
Cytoplasm
Cytoplasmic vesicle
Disulfide bond
Membrane
Metal-binding
Phosphoprotein
Reference proteome
Repeat
Synapse
Transmembrane
Transmembrane helix
Feature
chain Synaptotagmin-6
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
A0A3S2TLW8
A0A0N1IPK6
A0A212F3Z5
A0A0N1ID93
A0A1Y1N6J4
A0A067QR47
+ More
D1ZZC0 E2BB40 A0A158N9T6 A0A151JZB0 A0A195BI03 A0A151XI83 A0A3L8DRE0 A0A154PRL6 A0A195E5X8 A0A0J7KXR9 E2AP06 A0A026WGQ2 A0A232F3Z2 A0A195CIN9 F4WJX0 K7J3B0 A0A2A3EU03 A0A088AMX6 A0A1B6ECQ3 A0A0L7R5P7 A0A0M9A6E2 E0W2A7 A0A131YQL2 L7MFW0 A0A147BQ53 A0A1S3K2R7 A0A1S3K353 T1IQ10 K1PVV0 A0A2T7P5H7 A0A1E1XLK4 A0A433SRB2 V4CGA3 H3BFJ7 R7UL18 F6VZW5 G1KAX6 A0A3B5BB66 A0A151PGZ8 G5AVK1 H0V8W9 A0A091HTE3 C3ZW37 A0A2I2URS5 A0A2Y9NXN2 A0A2Y9H2P8 A0A2K5D6I0 A0A151PHW4 F6Q2I4 A0A3Q0FJB9 A0A3Q7S8N1 Q5T7P8-2 A0A2R9AZA0 I6L9C3 A0A340YID4 A0A1U7RM90 A0A1U7T955 A0A2Y9PBA5 A0A2K6V3T0 F7I632 A0A2Y9PBA8 A0A151NID4 A0A3Q7P2T0 G1TVA8 A0A2K5D6K5 A0A151PGX3 A0A1U7TIX3 A0A151NIC0 A0A2Y9SE66 A0A2Y9P395 A0A341D101 A0A2U4A1B3 A0A341D1Y1 A0A2U3V6N4 A0A3Q0E2D5 A0A3Q0FNE9 A0A0D9S689 M3WIR7 A0A341D148 G3SG95 A0A2K6CE36 A0A096N490 H9FQC4 A0A2K5NCK8 A0A452U9F1 D2H4J7 A0A0G2JW68 A0A1L8HE86 G3TB93 A0A2K6CE75 A0A2K5WUT0 A0A2K5YNS0
D1ZZC0 E2BB40 A0A158N9T6 A0A151JZB0 A0A195BI03 A0A151XI83 A0A3L8DRE0 A0A154PRL6 A0A195E5X8 A0A0J7KXR9 E2AP06 A0A026WGQ2 A0A232F3Z2 A0A195CIN9 F4WJX0 K7J3B0 A0A2A3EU03 A0A088AMX6 A0A1B6ECQ3 A0A0L7R5P7 A0A0M9A6E2 E0W2A7 A0A131YQL2 L7MFW0 A0A147BQ53 A0A1S3K2R7 A0A1S3K353 T1IQ10 K1PVV0 A0A2T7P5H7 A0A1E1XLK4 A0A433SRB2 V4CGA3 H3BFJ7 R7UL18 F6VZW5 G1KAX6 A0A3B5BB66 A0A151PGZ8 G5AVK1 H0V8W9 A0A091HTE3 C3ZW37 A0A2I2URS5 A0A2Y9NXN2 A0A2Y9H2P8 A0A2K5D6I0 A0A151PHW4 F6Q2I4 A0A3Q0FJB9 A0A3Q7S8N1 Q5T7P8-2 A0A2R9AZA0 I6L9C3 A0A340YID4 A0A1U7RM90 A0A1U7T955 A0A2Y9PBA5 A0A2K6V3T0 F7I632 A0A2Y9PBA8 A0A151NID4 A0A3Q7P2T0 G1TVA8 A0A2K5D6K5 A0A151PGX3 A0A1U7TIX3 A0A151NIC0 A0A2Y9SE66 A0A2Y9P395 A0A341D101 A0A2U4A1B3 A0A341D1Y1 A0A2U3V6N4 A0A3Q0E2D5 A0A3Q0FNE9 A0A0D9S689 M3WIR7 A0A341D148 G3SG95 A0A2K6CE36 A0A096N490 H9FQC4 A0A2K5NCK8 A0A452U9F1 D2H4J7 A0A0G2JW68 A0A1L8HE86 G3TB93 A0A2K6CE75 A0A2K5WUT0 A0A2K5YNS0
Pubmed
26354079
22118469
28004739
24845553
18362917
19820115
+ More
20798317 21347285 30249741 24508170 28648823 21719571 20075255 20566863 26830274 25576852 29652888 22992520 29209593 23254933 9215903 20431018 22293439 21993625 21993624 18563158 17975172 19892987 14702039 16710414 21406692 22722832 11181995 15489334 25243066 25319552 24813606 20010809 15057822 22673903 27762356
20798317 21347285 30249741 24508170 28648823 21719571 20075255 20566863 26830274 25576852 29652888 22992520 29209593 23254933 9215903 20431018 22293439 21993625 21993624 18563158 17975172 19892987 14702039 16710414 21406692 22722832 11181995 15489334 25243066 25319552 24813606 20010809 15057822 22673903 27762356
EMBL
RSAL01000059
RVE49665.1
KQ460426
KPJ14769.1
AGBW02010471
OWR48451.1
+ More
KQ459256 KPJ02074.1 GEZM01011427 JAV93524.1 KK853463 KDR07453.1 KQ971338 EFA01872.1 GL446901 EFN87131.1 ADTU01009516 ADTU01009517 ADTU01009518 ADTU01009519 KQ981404 KYN41947.1 KQ976476 KYM83837.1 KQ982093 KYQ60057.1 QOIP01000005 RLU22980.1 KQ435066 KZC14377.1 KQ979592 KYN20600.1 LBMM01002049 KMQ95352.1 GL441439 EFN64838.1 KK107214 EZA55242.1 NNAY01001071 OXU25249.1 KQ977791 KYM99928.1 GL888187 EGI65492.1 KZ288190 PBC34531.1 GEDC01001621 JAS35677.1 KQ414648 KOC66212.1 KQ435740 KOX76789.1 DS235875 EEB19763.1 GEDV01007679 JAP80878.1 GACK01002317 JAA62717.1 GEGO01002909 JAR92495.1 JH431273 JH818675 EKC25868.1 PZQS01000006 PVD28677.1 GFAA01003331 JAU00104.1 RQTK01001155 RUS71809.1 KB200559 ESP01100.1 AFYH01014035 AFYH01014036 AFYH01014037 AFYH01014038 AFYH01014039 AMQN01008151 KB302274 ELU04478.1 AAMC01049878 AAMC01049879 AAMC01049880 AKHW03000207 KYO48376.1 JH167162 EHB01062.1 AAKN02042499 AAKN02042500 AAKN02042501 KL217743 KFO98377.1 GG666695 EEN43251.1 AANG04003279 KYO48375.1 AK056448 AL162594 CH471122 AJFE02015061 AJFE02015062 AJFE02015063 BC044948 AAH44948.1 EAW56597.1 GAMS01008500 GAMS01008499 JAB14636.1 AKHW03002956 KYO36547.1 AAGW02000813 KYO48377.1 KYO36546.1 AQIB01086193 AANG04001751 CABD030005456 CABD030005457 AHZZ02016688 JU333078 AFE76833.1 GL192489 EFB24628.1 AABR07012755 AABR07012756 AABR07012757 CM004468 OCT94341.1 AQIA01002838 AQIA01002839 AQIA01002840 AQIA01002841
KQ459256 KPJ02074.1 GEZM01011427 JAV93524.1 KK853463 KDR07453.1 KQ971338 EFA01872.1 GL446901 EFN87131.1 ADTU01009516 ADTU01009517 ADTU01009518 ADTU01009519 KQ981404 KYN41947.1 KQ976476 KYM83837.1 KQ982093 KYQ60057.1 QOIP01000005 RLU22980.1 KQ435066 KZC14377.1 KQ979592 KYN20600.1 LBMM01002049 KMQ95352.1 GL441439 EFN64838.1 KK107214 EZA55242.1 NNAY01001071 OXU25249.1 KQ977791 KYM99928.1 GL888187 EGI65492.1 KZ288190 PBC34531.1 GEDC01001621 JAS35677.1 KQ414648 KOC66212.1 KQ435740 KOX76789.1 DS235875 EEB19763.1 GEDV01007679 JAP80878.1 GACK01002317 JAA62717.1 GEGO01002909 JAR92495.1 JH431273 JH818675 EKC25868.1 PZQS01000006 PVD28677.1 GFAA01003331 JAU00104.1 RQTK01001155 RUS71809.1 KB200559 ESP01100.1 AFYH01014035 AFYH01014036 AFYH01014037 AFYH01014038 AFYH01014039 AMQN01008151 KB302274 ELU04478.1 AAMC01049878 AAMC01049879 AAMC01049880 AKHW03000207 KYO48376.1 JH167162 EHB01062.1 AAKN02042499 AAKN02042500 AAKN02042501 KL217743 KFO98377.1 GG666695 EEN43251.1 AANG04003279 KYO48375.1 AK056448 AL162594 CH471122 AJFE02015061 AJFE02015062 AJFE02015063 BC044948 AAH44948.1 EAW56597.1 GAMS01008500 GAMS01008499 JAB14636.1 AKHW03002956 KYO36547.1 AAGW02000813 KYO48377.1 KYO36546.1 AQIB01086193 AANG04001751 CABD030005456 CABD030005457 AHZZ02016688 JU333078 AFE76833.1 GL192489 EFB24628.1 AABR07012755 AABR07012756 AABR07012757 CM004468 OCT94341.1 AQIA01002838 AQIA01002839 AQIA01002840 AQIA01002841
Proteomes
UP000283053
UP000053240
UP000007151
UP000053268
UP000027135
UP000007266
+ More
UP000008237 UP000005205 UP000078541 UP000078540 UP000075809 UP000279307 UP000076502 UP000078492 UP000036403 UP000000311 UP000053097 UP000215335 UP000078542 UP000007755 UP000002358 UP000242457 UP000005203 UP000053825 UP000053105 UP000009046 UP000085678 UP000005408 UP000245119 UP000271974 UP000030746 UP000008672 UP000014760 UP000008143 UP000001646 UP000261400 UP000050525 UP000006813 UP000005447 UP000054308 UP000001554 UP000011712 UP000248483 UP000248481 UP000233020 UP000002281 UP000189705 UP000286640 UP000005640 UP000240080 UP000265300 UP000189704 UP000233220 UP000008225 UP000286641 UP000001811 UP000248484 UP000252040 UP000245320 UP000029965 UP000001519 UP000233120 UP000028761 UP000233060 UP000291021 UP000002494 UP000186698 UP000007646 UP000233100 UP000233140
UP000008237 UP000005205 UP000078541 UP000078540 UP000075809 UP000279307 UP000076502 UP000078492 UP000036403 UP000000311 UP000053097 UP000215335 UP000078542 UP000007755 UP000002358 UP000242457 UP000005203 UP000053825 UP000053105 UP000009046 UP000085678 UP000005408 UP000245119 UP000271974 UP000030746 UP000008672 UP000014760 UP000008143 UP000001646 UP000261400 UP000050525 UP000006813 UP000005447 UP000054308 UP000001554 UP000011712 UP000248483 UP000248481 UP000233020 UP000002281 UP000189705 UP000286640 UP000005640 UP000240080 UP000265300 UP000189704 UP000233220 UP000008225 UP000286641 UP000001811 UP000248484 UP000252040 UP000245320 UP000029965 UP000001519 UP000233120 UP000028761 UP000233060 UP000291021 UP000002494 UP000186698 UP000007646 UP000233100 UP000233140
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A3S2TLW8
A0A0N1IPK6
A0A212F3Z5
A0A0N1ID93
A0A1Y1N6J4
A0A067QR47
+ More
D1ZZC0 E2BB40 A0A158N9T6 A0A151JZB0 A0A195BI03 A0A151XI83 A0A3L8DRE0 A0A154PRL6 A0A195E5X8 A0A0J7KXR9 E2AP06 A0A026WGQ2 A0A232F3Z2 A0A195CIN9 F4WJX0 K7J3B0 A0A2A3EU03 A0A088AMX6 A0A1B6ECQ3 A0A0L7R5P7 A0A0M9A6E2 E0W2A7 A0A131YQL2 L7MFW0 A0A147BQ53 A0A1S3K2R7 A0A1S3K353 T1IQ10 K1PVV0 A0A2T7P5H7 A0A1E1XLK4 A0A433SRB2 V4CGA3 H3BFJ7 R7UL18 F6VZW5 G1KAX6 A0A3B5BB66 A0A151PGZ8 G5AVK1 H0V8W9 A0A091HTE3 C3ZW37 A0A2I2URS5 A0A2Y9NXN2 A0A2Y9H2P8 A0A2K5D6I0 A0A151PHW4 F6Q2I4 A0A3Q0FJB9 A0A3Q7S8N1 Q5T7P8-2 A0A2R9AZA0 I6L9C3 A0A340YID4 A0A1U7RM90 A0A1U7T955 A0A2Y9PBA5 A0A2K6V3T0 F7I632 A0A2Y9PBA8 A0A151NID4 A0A3Q7P2T0 G1TVA8 A0A2K5D6K5 A0A151PGX3 A0A1U7TIX3 A0A151NIC0 A0A2Y9SE66 A0A2Y9P395 A0A341D101 A0A2U4A1B3 A0A341D1Y1 A0A2U3V6N4 A0A3Q0E2D5 A0A3Q0FNE9 A0A0D9S689 M3WIR7 A0A341D148 G3SG95 A0A2K6CE36 A0A096N490 H9FQC4 A0A2K5NCK8 A0A452U9F1 D2H4J7 A0A0G2JW68 A0A1L8HE86 G3TB93 A0A2K6CE75 A0A2K5WUT0 A0A2K5YNS0
D1ZZC0 E2BB40 A0A158N9T6 A0A151JZB0 A0A195BI03 A0A151XI83 A0A3L8DRE0 A0A154PRL6 A0A195E5X8 A0A0J7KXR9 E2AP06 A0A026WGQ2 A0A232F3Z2 A0A195CIN9 F4WJX0 K7J3B0 A0A2A3EU03 A0A088AMX6 A0A1B6ECQ3 A0A0L7R5P7 A0A0M9A6E2 E0W2A7 A0A131YQL2 L7MFW0 A0A147BQ53 A0A1S3K2R7 A0A1S3K353 T1IQ10 K1PVV0 A0A2T7P5H7 A0A1E1XLK4 A0A433SRB2 V4CGA3 H3BFJ7 R7UL18 F6VZW5 G1KAX6 A0A3B5BB66 A0A151PGZ8 G5AVK1 H0V8W9 A0A091HTE3 C3ZW37 A0A2I2URS5 A0A2Y9NXN2 A0A2Y9H2P8 A0A2K5D6I0 A0A151PHW4 F6Q2I4 A0A3Q0FJB9 A0A3Q7S8N1 Q5T7P8-2 A0A2R9AZA0 I6L9C3 A0A340YID4 A0A1U7RM90 A0A1U7T955 A0A2Y9PBA5 A0A2K6V3T0 F7I632 A0A2Y9PBA8 A0A151NID4 A0A3Q7P2T0 G1TVA8 A0A2K5D6K5 A0A151PGX3 A0A1U7TIX3 A0A151NIC0 A0A2Y9SE66 A0A2Y9P395 A0A341D101 A0A2U4A1B3 A0A341D1Y1 A0A2U3V6N4 A0A3Q0E2D5 A0A3Q0FNE9 A0A0D9S689 M3WIR7 A0A341D148 G3SG95 A0A2K6CE36 A0A096N490 H9FQC4 A0A2K5NCK8 A0A452U9F1 D2H4J7 A0A0G2JW68 A0A1L8HE86 G3TB93 A0A2K6CE75 A0A2K5WUT0 A0A2K5YNS0
PDB
3HN8
E-value=2.11226e-82,
Score=779
Ontologies
GO
GO:0016021
GO:0017158
GO:0019905
GO:0001786
GO:0070382
GO:0071277
GO:0017156
GO:0014059
GO:0005886
GO:0030276
GO:0005509
GO:0005544
GO:0000149
GO:0016192
GO:0050796
GO:0006887
GO:0016020
GO:0007340
GO:0048306
GO:0097038
GO:0042803
GO:0060478
GO:0045956
GO:0007268
GO:0005546
GO:0007608
GO:0046982
GO:0019898
GO:0030672
GO:0099525
GO:0005829
GO:0099699
GO:0030054
GO:0098978
GO:0098793
GO:0005515
GO:0045454
GO:0003676
GO:0006182
GO:0051603
GO:0030145
GO:0006412
GO:0016876
GO:0043039
PANTHER
Topology
Subcellular location
Membrane Localized predominantly to endoplasmic reticulum (ER) and/or Golgi-like perinuclear compartment (By similarity). With evidence from 5 publications.
Cytoplasm
Cytosol
Cell membrane
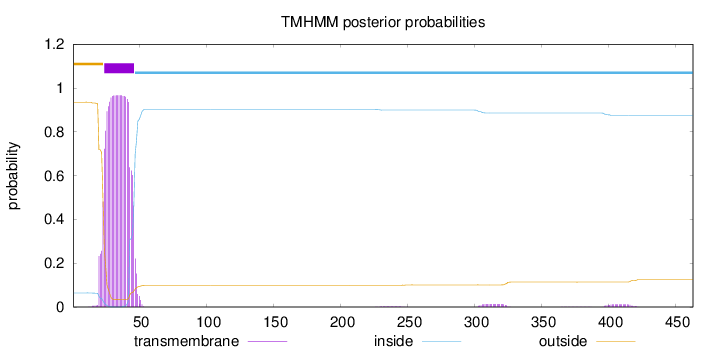
Length:
463
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.37495
Exp number, first 60 AAs:
21.82554
Total prob of N-in:
0.06445
POSSIBLE N-term signal
sequence
outside
1 - 23
TMhelix
24 - 46
inside
47 - 463
Population Genetic Test Statistics
Pi
224.05874
Theta
183.04711
Tajima's D
1.241228
CLR
0.271654
CSRT
0.719114044297785
Interpretation
Uncertain
